A diffusible cytotoxin of Haemophilus ducreyi
- PMID: 9108104
- PMCID: PMC20567
- DOI: 10.1073/pnas.94.8.4056
A diffusible cytotoxin of Haemophilus ducreyi
Abstract
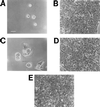
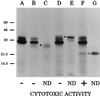
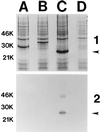
Little is known about the virulence mechanisms employed by Haemophilus ducreyi in the production of genital ulcers. This Gram-negative bacterium previously has been shown to produce a soluble cytotoxic activity that kills HeLa and HEp-2 cells. We have now identified a cluster of three H. ducreyi genes that encode this cytotoxic activity. The predicted proteins encoded by these genes are most similar to the products of the Escherichia coli cdtABC genes that comprise the cytolethal distending toxin (CDT) of this enteric pathogen. Eleven of 12 H. ducreyi strains were shown to possess this gene cluster and culture supernatants from these strains readily killed HeLa cells. The culture supernatant from a single strain of H. ducreyi that lacked these genes was unable to kill HeLa cells. When the H. ducreyi cdtABC gene cluster was cloned into E. coli, culture supernatant from the recombinant E. coli clone killed HeLa cells. A monoclonal antibody that neutralized this soluble cytotoxic activity of H. ducreyi was shown to bind to the H. ducreyi cdtC gene product. This soluble H. ducreyi cytotoxin may play a role in the development or persistence of the ulcerative lesions characteristic of chancroid.
Figures



References
-
- Ronald A R, Albritton W L. In: Sexually Transmitted Diseases. Holmes K K, Mardh P-A, Sparling P F, Wiesner P J, editors. New York: McGraw–Hill; 1990. pp. 263–271.
-
- Cameron D W, Simonsen J N, D’Costa L J, Ronald A R, Maitha G M, Gakinya M N, Cheang M, Piot P, Brunham R C, Plummer F A. Lancet. 1989;ii:403–408. - PubMed
-
- Plummer F A, Wainberg M A, Plourde P, Jessamine P G, D’Costa J L, Wamola I A, Ronald A R. J Infect Dis. 1990;161:810–811. - PubMed
-
- Odumeru J A, Wiseman G M, Ronald A R. J Med Microbiol. 1987;23:155–162. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

