Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage phi6
- PMID: 9108107
- PMCID: PMC20570
- DOI: 10.1073/pnas.94.8.4074
Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage phi6
Abstract
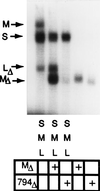
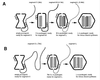
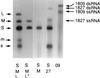
A model that explains the stoichiometric packaging of the chromosomes of phi6, a bacteriophage with a genome of three unique double-stranded RNA segments, is proposed and supported. Ordered switches in packaging specificity and RNA synthesis are determined by the amount of RNA within the procapsid. The plus strand of segment S binds to one of several sites on the outside of the empty procapsid. The RNA enters and the procapsid expands so that the S sites are lost and M sites appear. Packaging of segment M results in the loss of the M sites and the appearance of the L sites. Packaging of L readies the particle for minus-strand synthesis. If any of the segments is less than normal size, packaging of that class of segments continues until the normal content of RNA for that segment is packaged and the binding sites then change.
Figures




Similar articles
-
Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6.Microbiol Mol Biol Rev. 1999 Mar;63(1):149-60. doi: 10.1128/MMBR.63.1.149-160.1999. Microbiol Mol Biol Rev. 1999. PMID: 10066834 Free PMC article. Review.
-
In vivo studies of genomic packaging in the dsRNA bacteriophage Phi8.BMC Microbiol. 2005 Mar 11;5:10. doi: 10.1186/1471-2180-5-10. BMC Microbiol. 2005. PMID: 15762996 Free PMC article.
-
Packaging and replication regulation revealed by chimeric genome segments of double-stranded RNA bacteriophage phi6.RNA. 1999 Mar;5(3):446-54. doi: 10.1017/s1355838299981876. RNA. 1999. PMID: 10094312 Free PMC article.
-
Directed changes in the number of double-stranded RNA genomic segments in bacteriophage phi6.Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):3920-4. doi: 10.1073/pnas.95.7.3920. Proc Natl Acad Sci U S A. 1998. PMID: 9520468 Free PMC article.
-
Packaging in dsRNA viruses.Adv Exp Med Biol. 2012;726:601-8. doi: 10.1007/978-1-4614-0980-9_26. Adv Exp Med Biol. 2012. PMID: 22297532 Review.
Cited by
-
Genome packaging in multi-segmented dsRNA viruses: distinct mechanisms with similar outcomes.Curr Opin Virol. 2018 Dec;33:106-112. doi: 10.1016/j.coviro.2018.08.001. Epub 2018 Aug 23. Curr Opin Virol. 2018. PMID: 30145433 Free PMC article. Review.
-
Heterologous RNA Recombination in the Cystoviruses φ6 and φ8: A Mechanism of Viral Variation and Genome Repair.Viruses. 2022 Nov 21;14(11):2589. doi: 10.3390/v14112589. Viruses. 2022. PMID: 36423198 Free PMC article. Review.
-
Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage phi6 RNA-dependent RNA polymerase.RNA. 2007 Mar;13(3):422-9. doi: 10.1261/rna.348307. Epub 2007 Jan 19. RNA. 2007. PMID: 17237359 Free PMC article.
-
Analysis of specific binding involved in genomic packaging of the double-stranded-RNA bacteriophage phi6.J Bacteriol. 2003 Nov;185(21):6409-14. doi: 10.1128/JB.185.21.6409-6414.2003. J Bacteriol. 2003. PMID: 14563876 Free PMC article.
-
Further characterisation of rotavirus cores: Ss(+)RNAs can be packaged in vitro but packaging lacks sequence specificity.Virus Res. 2013 Dec 26;178(2):252-63. doi: 10.1016/j.virusres.2013.09.034. Epub 2013 Oct 1. Virus Res. 2013. PMID: 24091366 Free PMC article.
References
-
- Casjens S. In: Virus Structure and Assembly. Casjens S, editor. Boston: Jones & Bartlett; 1985. pp. 75–147.
-
- Black L W. In: The Bacteriophages. Calendar R, editor. Vol. 2. New York: Plenum; 1988. pp. 321–373.
-
- Tyler K L, Fields B N. In: Virology. Fields B N, Knipe D M, editors. New York: Raven; 1990. pp. 1271–1273.
-
- Spendlove R S, McClain M E, Lennette E H. J Gen Virol. 1970;8:83–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

