Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta
- PMID: 9113986
- PMCID: PMC20719
- DOI: 10.1073/pnas.94.9.4312
Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta
Abstract
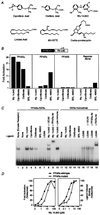
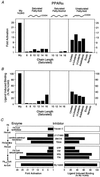
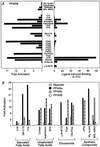
Fatty acids (FAs) and their derivatives are essential cellular metabolites whose concentrations must be closely regulated. This implies that regulatory circuits exist which can sense changes in FA levels. Indeed, the peroxisome proliferator-activated receptor alpha (PPARalpha) regulates lipid homeostasis and is transcriptionally activated by a variety of lipid-like compounds. It remains unclear as to how these structurally diverse compounds can activate a single receptor. We have developed a novel conformation-based assay that screens activators for their ability to bind to PPARalpha/delta and induce DNA binding. We show here that specific FAs, eicosanoids, and hypolipidemic drugs are ligands for PPARalpha or PPARdelta. Because altered FA levels are associated with obesity, atherosclerosis, hypertension, and diabetes, PPARs may serve as molecular sensors that are central to the development and treatment of these metabolic disorders.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

