Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma
- PMID: 9113987
- PMCID: PMC20720
- DOI: 10.1073/pnas.94.9.4318
Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma
Abstract
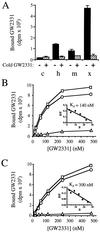
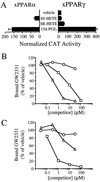
Peroxisome proliferator-activated receptors (PPARs) alpha and gamma are key regulators of lipid homeostasis and are activated by a structurally diverse group of compounds including fatty acids, eicosanoids, and hypolipidemic drugs such as fibrates and thiazolidinediones. While thiazolidinediones and 15-deoxy-Delta12, 14-prostaglandin J2 have been shown to bind to PPARgamma, it has remained unclear whether other activators mediate their effects through direct interactions with the PPARs or via indirect mechanisms. Here, we describe a novel fibrate, designated GW2331, that is a high-affinity ligand for both PPARalpha and PPARgamma. Using GW2331 as a radioligand in competition binding assays, we show that certain mono- and polyunsaturated fatty acids bind directly to PPARalpha and PPARgamma at physiological concentrations, and that the eicosanoids 8(S)-hydroxyeicosatetraenoic acid and 15-deoxy-Delta12,14-prostaglandin J2 can function as subtype-selective ligands for PPARalpha and PPARgamma, respectively. These data provide evidence that PPARs serve as physiological sensors of lipid levels and suggest a molecular mechanism whereby dietary fatty acids can modulate lipid homeostasis.
Figures



References
-
- Horrobin D F. Prostaglandins Leukotrienes Essent Fatty Acids. 1995;53:385–396. - PubMed
-
- Clarke S D, Jump D B. Lipids. 1996;37:S7–S11. - PubMed
-
- Hillgartner F B, Salati L M, Goodridge A G. Physiol Rev. 1995;75:47–76. - PubMed
-
- Amri E-Z, Ailhaud G, Grimaldi P-A. J Lipid Res. 1994;35:930–937. - PubMed
-
- Issemann I, Green S. Nature (London) 1990;347:645–650. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

