P110delta, a novel phosphoinositide 3-kinase in leukocytes
- PMID: 9113989
- PMCID: PMC20722
- DOI: 10.1073/pnas.94.9.4330
P110delta, a novel phosphoinositide 3-kinase in leukocytes
Abstract
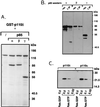
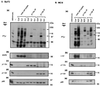
Phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases that have been implicated in signal transduction through tyrosine kinase- and heterotrimeric G-protein-linked receptors. We report herein the cloning and characterization of p110delta, a novel class I PI3K. Like p110alpha and p110beta, other class I PI3Ks, p110delta displays a broad phosphoinositide lipid substrate specificity and interacts with SH2/SH3 domain-containing p85 adaptor proteins and with GTP-bound Ras. In contrast to the widely distributed p110alpha and beta, p110delta is exclusively found in leukocytes. In these cells, p110alpha and delta both associate with the p85alpha and beta adaptor subunits and are similarly recruited to activated signaling complexes after treatment with the cytokines interleukin 3 and 4 and stem cell factor. Thus, these class I PI3Ks appear not to be distinguishable at the level of p85 adaptor selection or recruitment to activated receptor complexes. However, distinct biochemical and structural features of p110delta suggest divergent functional/regulatory capacities for this PI3K. Unlike p110alpha, p110delta does not phosphorylate p85 but instead harbors an intrinsic autophosphorylation capacity. In addition, the p110delta catalytic domain contains unique potential protein-protein interaction modules such as a Pro-rich region and a basic-region leucine-zipper (bZIP)-like domain. Possible selective functions of p110delta in white blood cells are discussed.
Figures



References
-
- Zvelebil M J, MacDougall L, Leevers S, Volinia S, Vanhaesebroeck B, Gout I, Panayotou G, Domin J, Stein R, Koga H, Salim K, Linacre J, Das P, Panaretou C, Wetzker R, Waterfield M D. Philos Trans R Soc London. 1996;351:217–233. - PubMed
-
- Stephens L R, Jackson T R, Hawkins P T. Biochim Biophys Acta. 1993;1179:27–75. - PubMed
-
- Fry M. Biochim Biophys Acta. 1994;1226:237–268. - PubMed
-
- Kapeller R, Cantley L C. BioEssays. 1994;8:565–576. - PubMed
-
- Vanhaesebroeck B, Stein R, Waterfield M D. Cancer Surv. 1996;27:249–270. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

