Carcinogens induce reversion of the mouse pink-eyed unstable mutation
- PMID: 9114032
- PMCID: PMC20765
- DOI: 10.1073/pnas.94.9.4576
Carcinogens induce reversion of the mouse pink-eyed unstable mutation
Abstract
Deletions and other genome rearrangements are associated with carcinogenesis and inheritable diseases. The pink-eyed unstable (pun) mutation in the mouse is caused by duplication of a 70-kb internal fragment of the p gene. Spontaneous reversion events in homozygous pun/pun mice occur through deletion of a duplicated sequence. Reversion events in premelanocytes in the mouse embryo detected as black spots on the gray fur of the offspring were inducible by the carcinogen x-rays, ethyl methanesulfonate, methyl methanesulfonate, ethyl nitrosourea, benzo[a]pyrene, trichloroethylene, benzene, and sodium arsenate. The latter three carcinogens are not detectable with several in vitro or in vivo mutagenesis assays. We studied the molecular mechanism of the carcinogen-induced reversion events by cDNA analysis using reverse transcriptase-PCR method and identified the induced reversion events as deletions. DNA deletion assays may be sensitive indicators for carcinogen exposure.
Figures

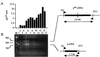
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

