The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain
- PMID: 9114061
- PMCID: PMC20794
- DOI: 10.1073/pnas.94.9.4739
The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain
Abstract
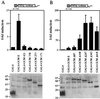
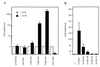
Absence or presence of glial cells missing (GCM) in cells of the developing nervous system of Drosophila decides over their future fate as neurons or glia with only those cells turning into glia that express GCM. To understand how GCM exerts its function we performed a detailed structure-function analysis. Using fusions between the DNA binding domain of the yeast GAL4 protein and GCM, we detected a transactivation function within the C-terminal part of GCM. In addition to this transactivation domain we mapped a sequence-specific DNA-binding domain within the N-terminal part of the GCM protein in close proximity to a bipartite nuclear localization signal. Binding site selection assays determined the motif 5'-AT(G/A)CGGGT-3' as the preferred binding site for GCM. Both the lack of homology to known proteins and the novel DNA binding specificity indicate that GCM contained a new type of DNA-binding domain. In transiently transfected cells, GCM also activated transcription from promoters consisting of the newly identified GCM-binding site and a TATA box. Thus, GCM is a novel type of transcription factor involved in early gliogenesis.
Figures
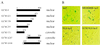
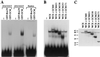
References
-
- Anderson D J. Neuron. 1989;3:1–12. - PubMed
-
- Doe C Q, Technau G M. Trends Neurosci. 1993;16:510–514. - PubMed
-
- Jan Y N, Jan L Y. Curr Opin Neurobiol. 1994;4:8–13. - PubMed
-
- Hosoya T, Takizawa K, Nitta K, Hotta Y. Cell. 1995;82:1025–1036. - PubMed
-
- Jones B W, Fetter R D, Tear G, Goodman C S. Cell. 1995;82:1013–1023. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

