A membrane-anchored E-type endo-1,4-beta-glucanase is localized on Golgi and plasma membranes of higher plants
- PMID: 9114071
- PMCID: PMC20804
- DOI: 10.1073/pnas.94.9.4794
A membrane-anchored E-type endo-1,4-beta-glucanase is localized on Golgi and plasma membranes of higher plants
Abstract
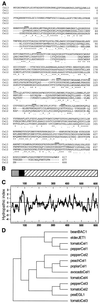
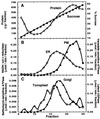
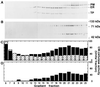
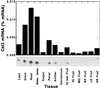
Endo-1,4-beta-D-glucanases (EGases, EC 3.2.1.4) are enzymes produced in bacteria, fungi, and plants that hydrolyze polysaccharides possessing a 1,4-beta-D-glucan backbone. All previously identified plant EGases are E-type endoglucanases that possess signal sequences for endoplasmic reticulum entry and are secreted to the cell wall. Here we report the characterization of a novel E-type plant EGase (tomato Cel3) with a hydrophobic transmembrane domain and structure typical of type II integral membrane proteins. The predicted protein is composed of 617 amino acids and possesses seven potential sites for N-glycosylation. Cel3 mRNA accumulates in young vegetative tissues with highest abundance during periods of rapid cell expansion, but is not hormonally regulated. Antibodies raised to a recombinant Cel3 protein specifically recognized three proteins, with apparent molecular masses of 93, 88, and 53 kDa, in tomato root microsomal membranes separated by sucrose density centrifugation. The 53-kDa protein comigrated in the gradient with plasma membrane markers, the 88-kDa protein with Golgi membrane markers, and the 93-kDa protein with markers for both Golgi and plasma membranes. EGase enzyme activity was also found in regions of the density gradient corresponding to both Golgi and plasma membranes, suggesting that Cel3 EGase resides in both membrane systems, the sites of cell wall polymer biosynthesis. The in vivo function of Cel3 is not known, but the only other known membrane-anchored EGase is present in Agrobacterium tumefaciens where it is required for cellulose biosynthesis.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

