NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses
- PMID: 9120401
- PMCID: PMC2196168
- DOI: 10.1084/jem.185.5.953
NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses
Abstract
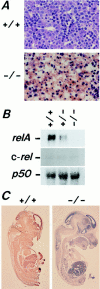
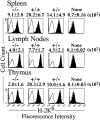
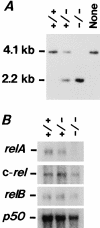
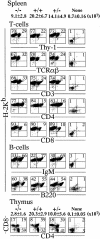
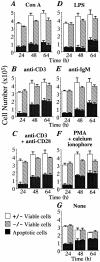
To investigate the function of NF-kappa B RelA (p65), we generated mice deficient in this NF-kappa B family member by homologous recombination. Mice lacking RelA showed liver degeneration and died around embryonic day 14.5. To elucidate the role of RelA in lymphocyte development and function, we transplanted fetal liver cells of 13.5-day embryos from heterozygote matings into irradiated SCID mice. Within 4 weeks, both T and B cells had developed in the SCID mice receiving relA-/- fetal liver transplants, similar to the relA+/+ and +/- cases. T cells were found to mature to Thy-1+/TCR alpha beta +/CD3+/CD4+ or CD8+, while B cells had the ability to differentiate to IgM+/B220+ and to secrete immunoglobulins. However, the secretion of IgG1 and IgA was reduced in RelA-deficient B cells. Furthermore, both T and B cells lacking RelA showed marked reduction in proliferative responses to stimulation with Con A, anti-CD3, anti-CD3 + anti-CD28, LPS, anti-IgM, and PMA + calcium ionophore. The results indicate that RelA plays a critical role in production of specific Ig isotypes and also in signal transduction pathways for lymphocyte proliferation.
Figures


References
-
- Baldwin AS., Jr The NF-κB and IκB proteins: New discoveries and insights. Annu Rev Immunol. 1996;14:649–681. - PubMed
-
- Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. - PubMed
-
- Baeuerle PA, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. - PubMed
-
- Verweij CL, Geerts M, Aarden LA. Activation of interleukin-2 gene transcription via T-cell surface molecule CD28 is mediated through an NF-κB-like response element. J Biol Chem. 1991;266:14179–14182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

