Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly
- PMID: 9122158
- PMCID: PMC20051
- DOI: 10.1073/pnas.94.6.2122
Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly
Abstract
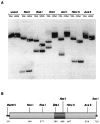
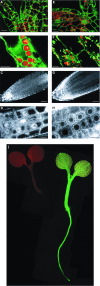
The green fluorescent protein (GFP) from the jellyfish Aequorea victoria is finding wide use as a genetic marker that can be directly visualized in the living cells of many heterologous organisms. We have sought to express GFP in the model plant Arabidopsis thaliana, but have found that proper expression of GFP is curtailed due to aberrant mRNA processing. An 84-nt cryptic intron is efficiently recognized and excised from transcripts of the GFP coding sequence. The cryptic intron contains sequences similar to those required for recognition of normal plant introns. We have modified the codon usage of the gfp gene to mutate the intron and to restore proper expression in Arabidopsis. GFP is mainly localized within the nucleoplasm and cytoplasm of transformed Arabidopsis cells and can give rise to high levels of fluorescence, but it proved difficult to efficiently regenerate transgenic plants from such highly fluorescent cells. However, when GFP is targeted to the endoplasmic reticulum, transformed cells regenerate routinely to give highly fluorescent plants. These modified forms of the gfp gene are useful for directly monitoring gene expression and protein localization and dynamics at high resolution, and as a simply scored genetic marker in living plants.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

