High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A
- PMID: 9128251
- PMCID: PMC2139767
- DOI: 10.1083/jcb.137.2.399
High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A
Erratum in
- J Cell Biol 1999 Sep 6;146(5):following 1201
Abstract
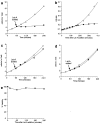
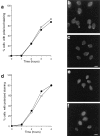
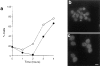
We report that the actin assembly inhibitor latrunculin-A (LAT-A) causes complete disruption of the yeast actin cytoskeleton within 2-5 min, suggesting that although yeast are nonmotile, their actin filaments undergo rapid cycles of assembly and disassembly in vivo. Differences in the LAT-A sensitivities of strains carrying mutations in components of the actin cytoskeleton suggest that tropomyosin, fimbrin, capping protein, Sla2p, and Srv2p act to increase actin cytoskeleton stability, while End3p and Sla1p act to decrease stability. Identification of three LAT-A resistant actin mutants demonstrated that in vivo effects of LAT-A are due specifically to impairment of actin function and implicated a region on the three-dimensional actin structure as the LAT-A binding site. LAT-A was used to determine which of 19 different proteins implicated in cell polarity development require actin to achieve polarized localization. Results show that at least two molecular pathways, one actin-dependent and the other actin-independent, underlie polarity development. The actin-dependent pathway localizes secretory vesicles and a putative vesicle docking complex to sites of cell surface growth, providing an explanation for the dependence of polarized cell surface growth on actin function. Unexpectedly, several proteins that function with actin during cell polarity development, including an unconventional myosin (Myo2p), calmodulin, and an actin-interacting protein (Bud6/Aip3p), achieved polarized localization by an actin-independent pathway, revealing interdependence among cell polarity pathways. Finally, transient actin depolymerization caused many cells to abandon one bud site or mating projection and to initiate growth at a second site. Thus, actin filaments are also required for maintenance of an axis of cell polarity.
Figures


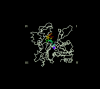









References
-
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nature Structural Biology. 1995;2:28–35. - PubMed
-
- Ayscough, K.R., and D.G. Drubin. 1997. Immunofluorescence microscopy of yeast cells. In Cell Biology: A Laboratory Handbook. 2nd Edition. J.E. Celis, Editor. Academic Press, San Diego, CA. In press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

