In vitro selection and evolution of functional proteins by using ribosome display
- PMID: 9144168
- PMCID: PMC24609
- DOI: 10.1073/pnas.94.10.4937
In vitro selection and evolution of functional proteins by using ribosome display
Abstract
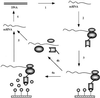
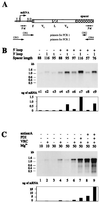
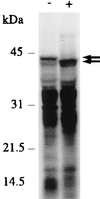
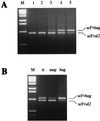
We report here a system with which a correctly folded complete protein and its encoding mRNA both remain attached to the ribosome and can be enriched for the ligand-binding properties of the native protein. We have selected a single-chain fragment (scFv) of an antibody 10(8)-fold by five cycles of transcription, translation, antigen-affinity selection, and PCR. The selected scFv fragments all mutated in vitro by acquiring up to four unrelated amino acid exchanges over the five generations, but they remained fully compatible with antigen binding. Libraries of native folded proteins can now be screened and made to evolve in a cell-free system without any transformation or constraints imposed by the host cell.
Figures
References
-
- Saffhill R, Schneider-Bernloehr H, Orgel L E, Spiegelman S. J Mol Biol. 1970;51:531–539. - PubMed
-
- Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. - PubMed
-
- Irvine D, Tuerk C, Gold L. J Mol Biol. 1991;222:739–761. - PubMed
-
- Dower W J, Cwirla S E. In: Guide to Electroporation and Electrofusion. Chang D C, Chassy B M, Saunders J A, Sowers A E, editors. San Diego: Academic; 1992. pp. 291–301.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

