Clustering of meiotic double-strand breaks on yeast chromosome III
- PMID: 9144217
- PMCID: PMC24658
- DOI: 10.1073/pnas.94.10.5213
Clustering of meiotic double-strand breaks on yeast chromosome III
Abstract
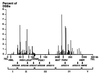
In the yeast Saccharomyces cerevisiae, meiotic recombination is initiated by transient DNA double-strand breaks (DSBs) that are repaired by interaction of the broken chromosome with its homologue. To identify a large number of DSB sites and gain insight into the control of DSB formation at both the local and the whole chromosomal levels, we have determined at high resolution the distribution of meiotic DSBs along the 340 kb of chromosome III. We have found 76 DSB regions, mostly located in intergenic promoter-containing intervals. The frequency of DSBs varies at least 50-fold from one region to another. The global distribution of DSB regions along chromosome III is nonrandom, defining large (39-105 kb) chromosomal domains, both hot and cold. The distribution of these localized DSBs indicates that they are likely to initiate most crossovers along chromosome III, but some discrepancies remain to be explained.
Figures
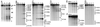

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

