A novel pair of immunoglobulin-like receptors expressed by B cells and myeloid cells
- PMID: 9144225
- PMCID: PMC24666
- DOI: 10.1073/pnas.94.10.5261
A novel pair of immunoglobulin-like receptors expressed by B cells and myeloid cells
Abstract
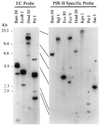
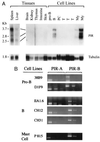
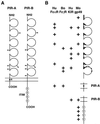
An Fcalpha receptor probe of human origin was used to identify novel members of the Ig gene superfamily in mice. Paired Ig-like receptors, named PIR-A and PIR-B, are predicted from sequence analysis of the cDNAs isolated from a mouse splenic library. Both type I transmembrane proteins possess similar ectodomains with six Ig-like loops, but have different transmembrane and cytoplasmic regions. The predicted PIR-A protein has a short cytoplasmic tail and a charged Arg residue in the transmembrane region that, by analogy with the FcalphaR relative, suggests the potential for association with an additional transmembrane protein to form a signal transducing unit. In contrast, the PIR-B protein has an uncharged transmembrane region and a long cytoplasmic tail containing four potential immunoreceptor tyrosine-based inhibitory motifs. These features are shared by the related killer inhibitory receptors. PIR-A proteins appear to be highly variable, in that predicted peptide sequences differ for seven randomly selected PIR-A clones, whereas PIR-B cDNA clones are invariant. Southern blot analysis with PIR-B and PIR-A-specific probes suggests only one PIR-B gene and multiple PIR-A genes. The PIR-A and PIR-B genes are expressed in B lymphocytes and myeloid lineage cells, wherein both are expressed simultaneously. The characteristics of the highly-conserved PIR-A and PIR-B genes and their coordinate cellular expression suggest a potential regulatory role in humoral, inflammatory, and allergic responses.
Figures


Comment in
-
Inhibitory receptors abound?Proc Natl Acad Sci U S A. 1997 Jun 10;94(12):5993-5. doi: 10.1073/pnas.94.12.5993. Proc Natl Acad Sci U S A. 1997. PMID: 9177155 Free PMC article. Review. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

