The Digital Anatomist distributed framework and its applications to knowledge-based medical imaging
- PMID: 9147337
- PMCID: PMC61233
- DOI: 10.1136/jamia.1997.0040165
The Digital Anatomist distributed framework and its applications to knowledge-based medical imaging
Abstract
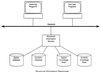
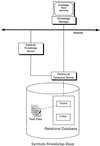
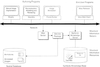
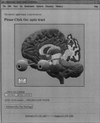
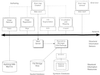
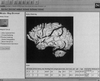
The domain of medical imaging is anatomy. Therefore, anatomic knowledge should be a rational basis for organizing and analyzing images. The goals of the Digital Anatomist Program at the University of Washington include the development of an anatomically based software framework for organizing, analyzing, visualizing and utilizing biomedical information. The framework is based on representations for both spatial and symbolic anatomic knowledge, and is being implemented in a distributed architecture in which multiple client programs on the Internet are used to update and access an expanding set of anatomical information resources. The development of this framework is driven by several practical applications, including symbolic anatomic reasoning, knowledge based image segmentation, anatomy information retrieval, and functional brain mapping. Since each of these areas involves many difficult image processing issues, our research strategy is an evolutionary one, in which applications are developed somewhat independently, and partial solutions are integrated in a piecemeal fashion, using the network as the substrate. This approach assumes that networks of interacting components can synergistically work together to solve problems larger than either could solve on its own. Each of the individual projects is described, along with evaluations that show that the individual components are solving the problems they were designed for, and are beginning to interact with each other in a synergistic manner. We argue that this synergy will increase, not only within our own group, but also among groups as the Internet matures, and that an anatomic knowledge base will be a useful means for fostering these interactions.
Figures




Comment in
-
Medical imaging informatics: challenges of definition and integration.J Am Med Inform Assoc. 1997 May-Jun;4(3):252-3. doi: 10.1136/jamia.1995.0040252. J Am Med Inform Assoc. 1997. PMID: 9147344 Free PMC article. No abstract available.
References
-
- Brinkley JF. Structural informatics and its applications in medicine and biology. Academic Medicine, 66: 589-91, 1991. - PubMed
-
- Rich E, Knight K. Artificial Intelligence. New York: McGraw-Hill, 2nd edition, 1991.
-
- Buchanan BG. Artificial intelligence as an experimental science. In: Aspects of Artificial Intelligence, Kluwer Academic Publishers, Dordrecht, Netherlands, 1988, pp. 209-50.
-
- Brinkley JF, Prothero JS, Prothero JW, Rosse C. A framework for the design of knowledge-based systems in structural biology. In 15th Annual Symposium on Computer Applications in Medical Care. New York: McGraw-Hill, 1989, pp. 61-5.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

