A proteasome cap subunit required for spindle pole body duplication in yeast
- PMID: 9151663
- PMCID: PMC2139890
- DOI: 10.1083/jcb.137.3.539
A proteasome cap subunit required for spindle pole body duplication in yeast
Abstract
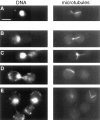
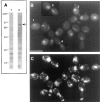
Proteasome-mediated protein degradation is a key regulatory mechanism in a diversity of complex processes, including the control of cell cycle progression. The selection of substrates for degradation clearly depends on the specificity of ubiquitination mechanisms, but further regulation may occur within the proteasomal 19S cap complexes, which attach to the ends of the 20S proteolytic core and are thought to control entry of substrates into the core. We have characterized a gene from Saccharomyces cerevisiae that displays extensive sequence similarity to members of a family of ATPases that are components of the 19S complex, including human subunit p42 and S. cerevisiae SUG1/CIM3 and CIM5 products. This gene, termed PCS1 (for proteasomal cap subunit), is identical to the recently described SUG2 gene (Russell, S.J., U.G. Sathyanarayana, and S.A. Johnston. 1996. J. Biol. Chem. 271:32810-32817). We have shown that PCS1 function is essential for viability. A temperature-sensitive pcs1 strain arrests principally in the second cycle after transfer to the restrictive temperature, blocking as large-budded cells with a G2 content of unsegregated DNA. EM reveals that each arrested pcs1 cell has failed to duplicate its spindle pole body (SPB), which becomes enlarged as in other monopolar mutants. Additionally, we have shown localization of a functional Pcs1-green fluorescent protein fusion to the nucleus throughout the cell cycle. We hypothesize that Pcs1p plays a role in the degradation of certain potentially nuclear component(s) in a manner that specifically is required for SPB duplication.
Figures




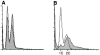

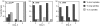

References
-
- Alber T. Structure of the leucine zipper. Curr Opin Gen Dev. 1992;2:205–210. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science (Wash DC) 1986;234:179–186. - PubMed
-
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

