Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis
- PMID: 9151691
- PMCID: PMC2139833
- DOI: 10.1083/jcb.137.4.891
Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis
Abstract
The gapA gene encoding a novel RasGTPase-activating protein (RasGAP)-related protein was found to be disrupted in a cytokinesis mutant of Dictyostelium that grows as giant and multinucleate cells in a dish culture. The predicted sequence of the GAPA protein showed considerable homology to those of Gap1/Sar1 from fission yeast and the COOH-terminal half of mammalian IQGAPs, the similarity extending beyond the RasGAP-related domain. In suspension culture, gapA- cells showed normal growth in terms of the increase in cell mass, but cytokinesis inefficiently occurred to produce spherical giant cells. Time-lapse recording of the dynamics of cell division in a dish culture revealed that, in the case of gapA- cells, cytokinesis was very frequently reversed at the step in which the midbody connecting the daughter cells should be severed. Earlier steps of cytokinesis in the gapA- cells seemed to be normal, since myosin II was accumulated at the cleavage furrow. Upon starvation, gapA- cells developed and formed fruiting bodies with viable spores, like the wild-type cells. These results indicate that the GAPA protein is specifically involved in the completion of cytokinesis. Recently, it was reported that IQGAPs are putative effectors for Rac and CDC42, members of the Rho family of GTPases, and participate in reorganization of the actin cytoskeleton. Thus, it is possible that Dictyostelium GAPA participates in the severing of the midbody by regulating the actin cytoskeleton through an interaction with a member of small GTPases.
Figures


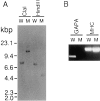





References
-
- Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideumcytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem Biophys Res Commun. 1994;205:1808–1814. - PubMed
-
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by rho-associated kinase (Rho-kinase) J Biol Chem. 1996a;271:20246–20249. - PubMed
-
- Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Identification of a putative target for Rho as the serine-threonine kinase protein kinase N. Science (Wash DC) 1996b;271:648–650. - PubMed
-
- Bain G, Tsang A. Disruption of the gene encoding the p34/31 polypeptides affects growth and development of Dictyostelium discoideum. . Mol Gen Genet. 1991;226:59–64. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

