Human macrophage-derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells
- PMID: 9151897
- PMCID: PMC2196293
- DOI: 10.1084/jem.185.9.1595
Human macrophage-derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells
Abstract
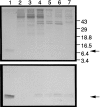
A cDNA encoding a novel human chemokine was isolated by random sequencing of cDNA clones from human monocyte-derived macrophages. This protein has been termed macrophage-derived chemokine (MDC) because it appears to be synthesized specifically by cells of the macrophage lineage. MDC has the four-cysteine motif and other highly conserved residues characteristic of CC chemokines, but it shares <35% identity with any of the known chemokines. Recombinant MDC was expressed in Chinese hamster ovary cells and purified by heparin-Sepharose chromatography. NH2-terminal sequencing and mass spectrophotometry were used to verify the NH2 terminus and molecular mass of recombinant MDC (8,081 dalton). In microchamber migration assays, monocyte-derived dendritic cells and IL-2-activated natural killer cells migrated to MDC in a dose-dependent manner, with a maximal chemotactic response at 1 ng/ml. Freshly isolated monocytes also migrated toward MDC, but with a peak response at 100 ng/ml MDC. Northern analyses indicated MDC is highly expressed in macrophages and in monocyte-derived dendritic cells, but not in monocytes, natural killer cells, or several cell lines of epithelial, endothelial, or fibroblast origin. High expression was also detected in normal thymus and less expression in lung and spleen. Unlike most other CC chemokines, MDC is encoded on human chromosome 16. MDC is thus a unique member of the CC chemokine family that may play a fundamental role in the function of dendritic cells, natural killer cells, and monocytes.
Figures










References
-
- Oppenheim JJ. Overview of chemokines. Adv Exp Med Biol. 1993;351:183–186. - PubMed
-
- Baggiolini M, Dewald B, Moser B. Interleukin-8 and related chemotactic cytokines-CXC and CC chemokines. Adv Immunol. 1994;55:97–179. - PubMed
-
- Schall TJ. Biology of the RANTES/SIS cytokine family. Cytokine. 1991;3:165–183. - PubMed
-
- Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science (Wash DC) 1993;261:600–603. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

