The second finger of Urbs1 is required for iron-mediated repression of sid1 in Ustilago maydis
- PMID: 9159169
- PMCID: PMC20875
- DOI: 10.1073/pnas.94.11.5882
The second finger of Urbs1 is required for iron-mediated repression of sid1 in Ustilago maydis
Abstract
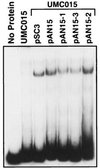
The urbs1 gene encodes a transcriptional regulator of siderophore biosynthesis in Ustilago maydis. Biological and DNA-binding activities of the two putative zinc-finger motifs of Urbs1 were studied by analyzing mutants containing altered finger domains. The mutated urbs1 alleles from three previously described N'-methyl-N'-nitro-N-nitrosoguanidine (NTG) mutants were mapped and cloned by a gap-repair procedure. Sequence analyses revealed single amino acid substitutions in two of the NTG mutants. Both mutations (G-507 to D in urbs1-1 and P-491 to L in urbs1-3), which are located in the Urbs1 C-terminal finger domain, reduced DNA-binding activity by 10-fold and were sufficient to confer a urbs1-minus phenotype. The third NTG urbs1 mutant (urbs1-2) also contained a mutation in one of the conserved amino acids (P-518 to S) in the C-terminal finger domain, but this mutation alone was not sufficient to confer a urbs1-minus phenotype. A second frame shift mutation was identified in urbs1-2 and is necessary for the urbs1-minus phenotype. In an analysis of the function of the N-terminal finger of Urbs1, the conserved amino acid Arg-350 was mutated to leucine. A Urbs1 protein with this mutation complemented a urbs1 null mutant strain. By contrast, a similar mutation in the C-terminal domain abolished the ability of Urbs1 to regulate siderophore biosynthesis and greatly reduced its ability to bind target DNA.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

