Expression of a divergent expansin gene is fruit-specific and ripening-regulated
- PMID: 9159182
- PMCID: PMC20888
- DOI: 10.1073/pnas.94.11.5955
Expression of a divergent expansin gene is fruit-specific and ripening-regulated
Abstract
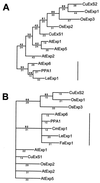
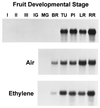
Expansins are proteins that induce extension in isolated plant cell walls in vitro and have been proposed to disrupt noncovalent interactions between hemicellulose and cellulose microfibrils. Because the plant primary cell wall acts as a constraint to cell enlargement, this process may be integral to plant cell expansion, and studies of expansins have focused on their role in growth. We report the identification of an expansin (LeExp1) from tomato that exhibits high levels of mRNA abundance and is specifically expressed in ripening fruit, a developmental period when growth has ceased but when selective disassembly of cell wall components is pronounced. cDNAs closely related to LeExp1 were also identified in ripening melons and strawberries, suggesting that they are a common feature of fruit undergoing rapid softening. Furthermore, the sequence of LeExp1 and its homologs from other ripening fruit define a subclass of expansin genes. Expression of LeExp1 is regulated by ethylene, a hormone known to coordinate and induce ripening in many species. LeExp1 is differentially expressed in the ripening-impaired tomato mutants Nr, rin, and nor, and mRNA abundance appears to be influenced directly by ethylene and by a developmentally modulated transduction pathway. The identification of a ripening-regulated expansin gene in tomato and other fruit suggests that, in addition to their role in facilitating the expansion of plant cells, expansins may also contribute to cell wall disassembly in nongrowing tissues, possibly by enhancing the accessibility of noncovalently bound polymers to endogenous enzymic action.
Figures

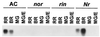



Comment in
-
Creeping walls, softening fruit, and penetrating pollen tubes: the growing roles of expansins.Proc Natl Acad Sci U S A. 1997 May 27;94(11):5504-5. doi: 10.1073/pnas.94.11.5504. Proc Natl Acad Sci U S A. 1997. PMID: 9159099 Free PMC article. Review. No abstract available.
References
-
- Carpita N C, Gibeaut D M. Plant J. 1993;3:1–30. - PubMed
-
- McCann M C, Wells B, Roberts K. J Cell Sci. 1990;96:323–334.
-
- Fry S C. Physiol Plant. 1989;75:532–536.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

