Discovery of a second 15S-lipoxygenase in humans
- PMID: 9177185
- PMCID: PMC21017
- DOI: 10.1073/pnas.94.12.6148
Discovery of a second 15S-lipoxygenase in humans
Abstract
The lipoxygenase metabolism of arachidonic acid occurs in specific blood cell types and epithelial tissues and is activated in inflammation and tissue injury. In the course of studying lipoxygenase expression in human skin, we detected and characterized a previously unrecognized enzyme that at least partly accounts for the 15S-lipoxygenase metabolism of arachidonic acid in certain epithelial tissues. The cDNA was cloned from human hair roots, and expression of the mRNA was detected also in prostate, lung, and cornea; an additional 16 human tissues, including peripheral blood leukocytes, were negative for the mRNA. The cDNA encodes a protein of 676 amino acids with a calculated molecular mass of 76 kDa. The amino acid sequence has approximately 40% identity to the known human 5S-, 12S-, and 15S-lipoxygenases. When expressed in HEK 293 cells, the newly discovered enzyme converts arachidonic acid exclusively to 15S-hydroperoxyeicosatetraenoic acid, while linoleic acid is less well metabolized. These features contrast with the previously reported 15S-lipoxygenase, which oxygenates arachidonic acid mainly at C-15, but also partly at C-12, and for which linoleic acid is an excellent substrate. The different catalytic activities and tissue distribution suggest a distinct function for the new enzyme compared with the previously reported human 15S-lipoxygenase.
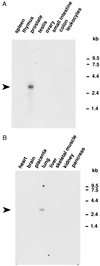
Figures



References
-
- Funk C D. Prog Nucleic Acid Res Mol Biol. 1993;45:67–98. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

