D-Ala-D-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB
- PMID: 9177243
- PMCID: PMC21075
- DOI: 10.1073/pnas.94.12.6480
D-Ala-D-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB
Abstract
The crisis in antibiotic resistance has resulted in an increasing fear of the emergence of untreatable organisms. Resistance to the glycopeptide antibiotic vancomycin in the enterococci, and the spread of these pathogens throughout the environment, has shown that this scenario is a matter of fact rather than fiction. The basis for vancomycin resistance is the manufacture of the depsipeptide D-Ala-D-lactate, which is incorporated into the peptidoglycan cell wall in place of the vancomycin target D-Ala-D-Ala. Pivotal to the resistance mechanism is the production of a D-Ala-D-Ala ligase capable of ester formation. Two highly efficient depsipeptide ligases have been cloned from vancomycin-resistant enterococci: VanA and VanB. These ligases show high amino acid sequence similarity to each other ( approximately 75%), but less so to other D-Ala-D-X ligases (<30%). We have cloned ddls from two glycopeptide-producing organisms, the vancomycin producer Amycolatopsis orientalis and the A47934 producer Streptomyces toyocaensis. These ligases show strong predicted amino acid homology to VanA and VanB (>60%) but not to other D-Ala-D-X ligases (<35%). The D-Ala-D-Ala ligase from S. toyocaensis shows D-Ala-D-lactate synthase activity in cell-free extracts of S. lividans transformed with the ddl gene and confirms the predicted enzymatic activity. These results imply a close evolutionary relationship between resistance mechanisms in the clinics and in drug-producing bacteria.
Figures





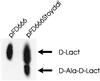
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

