Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle
- PMID: 9182667
- PMCID: PMC2132527
- DOI: 10.1083/jcb.137.6.1355
Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle
Abstract
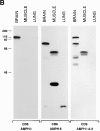
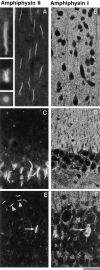
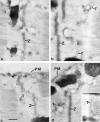
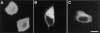
Amphiphysin (amphiphysin I), a dominant autoantigen in paraneoplastic Stiff-man syndrome, is a neuronal protein highly concentrated in nerve terminals, where it has a putative role in endocytosis. The yeast homologue of amphiphysin, Rvs167, has pleiotropic functions, including a role in endocytosis and in actin dynamics, suggesting that amphiphysin may also be implicated in the function of the presynaptic actin cytoskeleton. We report here the characterization of a second mammalian amphiphysin gene, amphiphysin II (SH3P9; BIN1), which encodes products primarily expressed in skeletal muscle and brain, as differentially spliced isoforms. In skeletal muscle, amphiphysin II is concentrated around T tubules, while in brain it is concentrated in the cytomatrix beneath the plasmamembrane of axon initial segments and nodes of Ranvier. In both these locations, amphiphysin II is colocalized with splice variants of ankyrin3 (ankyrinG), a component of the actin cytomatrix. In the same regions, the presence of clathrin has been reported. These findings support the hypothesis that, even in mammalian cells, amphiphysin/Rvs family members have a role both in endocytosis and in actin function and suggest that distinct amphiphysin isoforms contribute to define distinct domains of the cortical cytoplasm. Since amphiphysin II (BIN1) was reported to interact with Myc, it may also be implicated in a signaling pathway linking the cortical cytoplasm to nuclear function.
Figures









References
-
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. - PubMed
-
- Bogue CW, Gross I, Vasavada H, Dynia DW, Wilson CM, Jacobs HD. Identification of Hox genes in newborn lung and effects of gestational age and retinoic acid on their expression. Am J Physiol. 1994;266:L448–454. - PubMed
-
- Campos-Ortega JA, Glees P, Neuhoff V. Ultrastructural analysis of individual layers in the lateral geniculate body of the monkey. Z Zellforsch Mikrosk Anat. 1968;87:82–100. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

