A family of delayed rectifier Kv1 cDNAs showing cell type-specific expression in the squid stellate ganglion/giant fiber lobe complex
- PMID: 9185544
- PMCID: PMC6573319
- DOI: 10.1523/JNEUROSCI.17-13-05070.1997
A family of delayed rectifier Kv1 cDNAs showing cell type-specific expression in the squid stellate ganglion/giant fiber lobe complex
Abstract
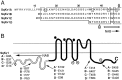
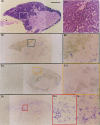
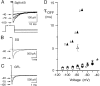
Squid giant axons are formed by giant fiber lobe (GFL) neurons of the stellate ganglion (SG). Other large motoneurons in the SG form a parallel system. A small family of cDNAs (SqKv1A-D) encoding Kv1 alpha-subunits was identified in a squid (Loligo opalescens) SG/GFL library. Members have distinct 5' untranslated regions (UTRs) and initial coding regions, but beyond a certain point (nucleotide 34 of SqKv1A) only nine differences exist. 3' UTRs are identical. Predicted alpha-subunits are nearly identical, and only the N termini differ significantly, primarily in length. RNase protection assays that use RNA isolated from specific SG regions show that SqKv1A mRNA is expressed prominently in the GFL but not in the SG proper. SqKv1B yields the opposite pattern. SqKv1D also is expressed only in the SG. SqKv1C expression was not detectable. In situ hybridizations confirm these results and reveal that SqKv1B mRNA is abundant in many large neurons of the SG, whereas SqKv1D expression is limited to small isolated clusters of neurons. SqKv1A and B are thus the predominant Kv1 mRNAs in the SG/GFL complex. Activation properties of SqKv1A and B channels expressed in oocytes are very similar to one another and compare favorably with properties of native delayed rectifier channels in GFL neurons and large SG neurons. The Kv1 complement in these squid neurons thus seems to be relatively simple. Several differences exist between cloned and native channels, however, and may reflect differences in the cellular environments of oocytes and neurons.
Figures



Similar articles
-
Inactivation and pharmacological properties of sqKv1A homotetramers in Xenopus oocytes cannot account for behavior of the squid "delayed rectifier" K(+) conductance.Biophys J. 2002 Jun;82(6):3022-36. doi: 10.1016/S0006-3495(02)75643-9. Biophys J. 2002. PMID: 12023225 Free PMC article.
-
Temperature-dependent expression of a squid Kv1 channel in Sf9 cells and functional comparison with the native delayed rectifier.J Membr Biol. 2001 Mar 15;180(2):147-61. doi: 10.1007/s002320010066. J Membr Biol. 2001. PMID: 11318098
-
Natural substitutions at highly conserved T1-domain residues perturb processing and functional expression of squid Kv1 channels.J Neurophysiol. 2001 Jan;85(1):61-71. doi: 10.1152/jn.2001.85.1.61. J Neurophysiol. 2001. PMID: 11152706
-
Molecular identification of SqKv1A. A candidate for the delayed rectifier K channel in squid giant axon.J Gen Physiol. 1996 Sep;108(3):207-19. doi: 10.1085/jgp.108.3.207. J Gen Physiol. 1996. PMID: 8882864 Free PMC article.
-
Spatial localization of calcium channels in giant fiber lobe neurons of the squid (Loligo opalescens).Proc Natl Acad Sci U S A. 1996 May 14;93(10):5067-71. doi: 10.1073/pnas.93.10.5067. Proc Natl Acad Sci U S A. 1996. PMID: 8643530 Free PMC article.
Cited by
-
Inactivation and pharmacological properties of sqKv1A homotetramers in Xenopus oocytes cannot account for behavior of the squid "delayed rectifier" K(+) conductance.Biophys J. 2002 Jun;82(6):3022-36. doi: 10.1016/S0006-3495(02)75643-9. Biophys J. 2002. PMID: 12023225 Free PMC article.
-
Voltage-gated potassium channels and the diversity of electrical signalling.J Physiol. 2012 Jun 1;590(11):2591-9. doi: 10.1113/jphysiol.2011.224212. Epub 2012 Mar 19. J Physiol. 2012. PMID: 22431339 Free PMC article. Review.
-
Ionic channel function in action potential generation: current perspective.Mol Neurobiol. 2007 Apr;35(2):129-50. doi: 10.1007/s12035-007-8001-0. Mol Neurobiol. 2007. PMID: 17917103 Review.
-
Molecular underpinnings of motor pattern generation: differential targeting of shal and shaker in the pyloric motor system.J Neurosci. 2000 Sep 1;20(17):6619-30. doi: 10.1523/JNEUROSCI.20-17-06619.2000. J Neurosci. 2000. PMID: 10964967 Free PMC article.
-
The emerging role of RNA editing in plasticity.J Exp Biol. 2015 Jun;218(Pt 12):1812-21. doi: 10.1242/jeb.119065. J Exp Biol. 2015. PMID: 26085659 Free PMC article. Review.
References
-
- Attali B, Lesage F, Ziliani P, Guillemare E, Honore E, Waldmann R, Hugnot JP, Mattei MG, Lazdunski M, Barhanin J. Multiple mRNA isoforms encoding the mouse cardiac Kv1–5 delayed rectifier K+ channel. J Biol Chem. 1993;268:24283–24289. - PubMed
-
- Dalbey RE, Von Heinje G. Signal peptidases in prokaryotes and eukaryotes: a new protease family. Trends Biochem Sci. 1992;17:474–478. - PubMed
-
- Gilly WF, Lucero MT, Horrigan FH. Control of the spatial distribution of sodium channels in giant fiber lobe neurons of the squid. Neuron. 1990;5:663–674. - PubMed
-
- Gilly WF, Hopkins B, Mackie GO. Development of giant motor axons and neural control of escape responses in squid embryos and hatchlings. Biol Bull. 1991;180:209–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
