Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors
- PMID: 9192616
- PMCID: PMC21209
- DOI: 10.1073/pnas.94.13.6629
Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors
Abstract
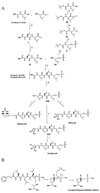
The proteasome is a multicatalytic protease complex that plays a key role in diverse cellular functions. The peptide vinyl sulfone, carboxybenzyl-leucyl-leucyl-leucine vinyl sulfone (Z-L3VS) covalently inhibits the trypsin-like, chymotrypsin-like and, unlike lactacystin, also the peptidylglutamyl peptidase activity in isolated proteasomes, and blocks their function in living cells. Although described as a class of mechanism-based inhibitors for cysteine proteases, the peptide vinyl sulfone Z-L3VS and a 125I-labeled nitrophenol derivative (125I-NIP-L3VS) covalently modify the active site threonine of the catalytic beta subunits of the proteasome. Modification of Thermoplasma proteasomes demonstrates the requirement for a hydroxyl amino acid (threonine, serine) as nucleophile at the beta subunit's NH2 terminus. 125I-NIP-L3VS covalently modifies the HslV subunit of the Escherichia coli protease complex HslV/HslU, a reaction that requires ATP, and supports a catalytic mechanism shared with that of the eukaryotic proteasome.
Figures



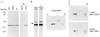


References
-
- Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. - PubMed
-
- Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. - PubMed
-
- Dahlmann B, Kopp F, Kuehn L, Niedel B, Pfeifer G, Hegerl R, Baumeister W. FEBS Lett. 1989;251:125–131. - PubMed
-
- Hershko A, Ciechanover A. Annu Rev Biochem. 1992;61:761–807. - PubMed
-
- Gaczynska M, Rock K L, Goldberg A L. Nature (London) 1993;365:264–267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

