RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein
- PMID: 9192668
- PMCID: PMC21261
- DOI: 10.1073/pnas.94.13.6927
RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein
Abstract
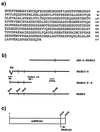
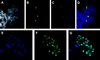
The human RAD51 protein is a homologue of the bacteria RecA and yeast RAD51 proteins that are involved in homologous recombination and DNA repair. RAD51 interacts with proteins involved in recombination and also with tumor suppressor proteins p53 and breast cancer susceptibility gene 1 (BRCA1). We have used the yeast two-hybrid system to clone murine cDNA sequences that encode two RAD51-associated molecules, RAB22 and RAB163. RAB163 encodes the C-terminal portion of mouse BRCA2, the homologue of the second breast cancer susceptibility gene protein in humans, demonstrating an in vitro association between RAD51 and BRCA2. RAB22 is a novel gene product that also interacts with RAD51 in vitro. To detect RAD51 interactions in vivo, we developed a transient nuclear focus assay that was used to demonstrate a complete colocalization of RAB22 with RAD51 in large nuclear foci.
Figures

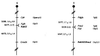

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

