Induced expression of trimerized intracellular domains of the human tumor necrosis factor (TNF) p55 receptor elicits TNF effects
- PMID: 9199176
- PMCID: PMC2137820
- DOI: 10.1083/jcb.137.7.1627
Induced expression of trimerized intracellular domains of the human tumor necrosis factor (TNF) p55 receptor elicits TNF effects
Abstract
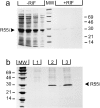
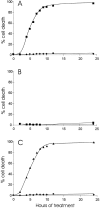
The various biological activities of tumor necrosis factor (TNF) are mediated by two receptors, one of 55 kD (TNF-R55) and one of 75 kD (TNF-R75). Although the phenotypic and molecular responses elicited by TNF in different cell types are fairly well characterized, the signaling pathways leading to them are so far only partly understood. To further unravel these processes, we focused on TNF-R55, which is responsible for mediating most of the known TNF effects. Since several studies have demonstrated the importance of receptor clustering and consequently of close association of the intracellular domains for signaling, we addressed the question of whether clustering of the intracellular domains of TNF-R55 (TNF-R55i) needs to occur in structural association with the inner side of the cell membrane, where many signaling mediators are known to reside. Therefore, we investigated whether induced intracellular clustering of only TNF-R55i would be sufficient to initiate and generate a full TNF response, without the need for a full-length receptor molecule or a transmembrane region. Our results provide clear evidence that inducible forced trimerization of either TNF-R55i or only the death domain elicits an efficient TNF response, comprising activation of the nuclear factor kappaB, induction of interleukin-6, and cell killing.
Figures







References
-
- Adam D, Keßler U, Krönke M. Cross-linking of the p55 tumor necrosis factor receptor cytoplasmic domain by a dimeric ligand induces nuclear factor-κB and mediates cell death. J Biol Chem. 1995;270:17482–17487. - PubMed
-
- Adam D, Adam-Klages S, Krönke M. Identification of p55 tumor necrosis factor receptor associated proteins that couple to signaling pathways not initiated by the death domain. J Inflamm. 1996a;47:61–66. - PubMed
-
- Adam D, Wiegmann K, Adam-Klages S, Ruff A, Krönke M. A novel cytoplasmic domain of the p55 TNF receptor initiates the neutral sphingomyelinase pathway. J Biol Chem. 1996b;271:14617–14622. - PubMed
-
- Adam-Klages S, Adam D, Wiegmann K, Struve S, Kolanus W, Schneider-Mergener J, Krönke M. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell. 1996;86:937–947. - PubMed
-
- Aggarwal BB, Natarajan K. Tumor necrosis factors: developments during the last decade. Eur Cytokine Netw. 1996;7:93–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

