Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria
- PMID: 9204897
- PMCID: PMC12235523
- DOI: 10.1126/science.277.5322.60
Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria
Erratum in
- Science 1997 Dec 19;278(5346):2037
Abstract
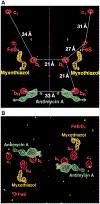
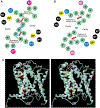
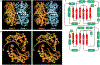
On the basis of x-ray diffraction data to a resolution of 2.9 angstroms, atomic models of most protein components of the bovine cytochrome bc1 complex were built, including core 1, core 2, cytochrome b, subunit 6, subunit 7, a carboxyl-terminal fragment of cytochrome c1, and an amino-terminal fragment of the iron-sulfur protein. The positions of the four iron centers within the bc1 complex and the binding sites of the two specific respiratory inhibitors antimycin A and myxothiazol were identified. The membrane-spanning region of each bc1 complex monomer consists of 13 transmembrane helices, eight of which belong to cytochrome b. Closely interacting monomers are arranged as symmetric dimers and form cavities through which the inhibitor binding pockets can be accessed. The proteins core 1 and core 2 are structurally similar to each other and consist of two domains of roughly equal size and identical folding topology.
Figures


References
-
- Schägger H, Link TA, Engel WD, Von Jagow G, Methods Enzymol 126, 224 (1986). - PubMed
-
- Gonzalez-Halphen D, Lindorfer MA, Capaldi RA, Biochemistry 27, 7021 (1988). - PubMed
-
- Wakabayashi S, Takeda H, Matsubara H, Kim CH, King TE, J. Biochem. (Tokyo) 91, 2077 (1982); - PubMed
- Wakabayashi S et al. , J. Biol. Chem 260, 337 (1985); - PubMed
- Schägger H, Borchart U, Aquila H, Link TA, Von Jagow G, FEBS Lett 190, 89 (1985); - PubMed
- Schägger H, Borchart U, Machleidt W, Link TA, Von Jagow G, ibid. 219, 161 (1987); - PubMed
- Borchart U, Machleidt W, Schägger H, Link TA, Von Jagow G, ibid. 191, 125 (1985); - PubMed
- ibid. 200, 81 (1986).
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

