Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein
- PMID: 9207077
- PMCID: PMC23807
- DOI: 10.1073/pnas.94.14.7251
Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein
Abstract
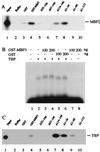
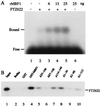
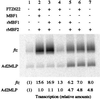
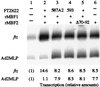
Multiprotein bridging factor 1 (MBF1) is a transcriptional cofactor that bridges between the TATA box-binding protein (TBP) and the Drosophila melanogaster nuclear hormone receptor FTZ-F1 or its silkworm counterpart BmFTZ-F1. A cDNA clone encoding MBF1 was isolated from the silkworm Bombyx mori whose sequence predicts a basic protein consisting of 146 amino acids. Bacterially expressed recombinant MBF1 is functional in interactions with TBP and a positive cofactor MBF2. The recombinant MBF1 also makes a direct contact with FTZ-F1 through the C-terminal region of the FTZ-F1 DNA-binding domain and stimulates the FTZ-F1 binding to its recognition site. The central region of MBF1 (residues 35-113) is essential for the binding of FTZ-F1, MBF2, and TBP. When the recombinant MBF1 was added to a HeLa cell nuclear extract in the presence of MBF2 and FTZ622 bearing the FTZ-F1 DNA-binding domain, it supported selective transcriptional activation of the fushi tarazu gene as natural MBF1 did. Mutations disrupting the binding of FTZ622 to DNA or MBF1, or a MBF2 mutation disrupting the binding to MBF1, all abolished the selective activation of transcription. These results suggest that tethering of the positive cofactor MBF2 to a FTZ-F1-binding site through FTZ-F1 and MBF1 is essential for the binding site-dependent activation of transcription. A homology search in the databases revealed that the deduced amino acid sequence of MBF1 is conserved across species from yeast to human.
Figures


References
-
- Johnson P, McKnight S. Annu Rev Biochem. 1989;58:799–839. - PubMed
-
- Mitchell P J, Tjian R. Science. 1989;245:371–378. - PubMed
-
- Lewin B. Cell. 1990;61:1161–1164. - PubMed
-
- Roeder R G. Trends Biochem Sci. 1991;16:402–408. - PubMed
-
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lim S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

