A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization
- PMID: 9214378
- PMCID: PMC2139937
- DOI: 10.1083/jcb.138.1.17
A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization
Abstract
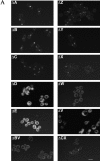
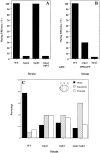
SPA2 encodes a yeast protein that is one of the first proteins to localize to sites of polarized growth, such as the shmoo tip and the incipient bud. The dynamics and requirements for Spa2p localization in living cells are examined using Spa2p green fluorescent protein fusions. Spa2p localizes to one edge of unbudded cells and subsequently is observable in the bud tip. Finally, during cytokinesis Spa2p is present as a ring at the mother-daughter bud neck. The bud emergence mutants bem1 and bem2 and mutants defective in the septins do not affect Spa2p localization to the bud tip. Strikingly, a small domain of Spa2p comprised of 150 amino acids is necessary and sufficient for localization to sites of polarized growth. This localization domain and the amino terminus of Spa2p are essential for its function in mating. Searching the yeast genome database revealed a previously uncharacterized protein which we name, Sph1p (a2p omolog), with significant homology to the localization domain and amino terminus of Spa2p. This protein also localizes to sites of polarized growth in budding and mating cells. SPH1, which is similar to SPA2, is required for bipolar budding and plays a role in shmoo formation. Overexpression of either Spa2p or Sph1p can block the localization of either protein fused to green fluorescent protein, suggesting that both Spa2p and Sph1p bind to and are localized by the same component. The identification of a 150-amino acid domain necessary and sufficient for localization of Spa2p to sites of polarized growth and the existence of this domain in another yeast protein Sph1p suggest that the early localization of these proteins may be mediated by a receptor that recognizes this small domain.
Figures











References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

