Control of mitotic events by Nap1 and the Gin4 kinase
- PMID: 9214386
- PMCID: PMC2139941
- DOI: 10.1083/jcb.138.1.119
Control of mitotic events by Nap1 and the Gin4 kinase
Abstract
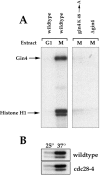
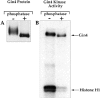
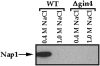
Little is known about the pathways used by cyclins and cyclin-dependent kinases to induce the events of the cell cycle. In budding yeast, a protein called Nap1 binds to the mitotic cyclin Clb2, and Nap1 is required for the ability of Clb2 to induce specific mitotic events, but the role played by Nap1 is unclear. We have used genetic and biochemical approaches to identify additional proteins that function with Nap1 in the control of mitotic events. These approaches have both identified a protein kinase called Gin4 that is required for the ability of Clb2 and Nap1 to promote the switch from polar to isotropic bud growth that normally occurs during mitosis. Gin4 is also required for the ability of Clb2 and Nap1 to promote normal progression through mitosis. The Gin4 protein becomes phosphorylated as cells enter mitosis, resulting in the activation of Gin4 kinase activity, and the phosphorylation of Gin4 is dependent upon Nap1 and Clb2 in vivo. Affinity chromatography experiments demonstrate that Gin4 binds tightly to Nap1, indicating that the functions of these two proteins are closely tied within the cell. These results demonstrate that the activation of Gin4 is under the control of Clb2 and Nap1, and they provide an important step towards elucidating the molecular pathways that link cyclin-dependent kinases to the events they control.
Figures








References
-
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. - PubMed
-
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. - PubMed
-
- Ghiara JB, Richardson HE, Sugimoto K, Henze M, Lew DJ, Witenberg C, Reed SI. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991;65:163–174. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

