Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits
- PMID: 9221782
- PMCID: PMC6573193
- DOI: 10.1523/JNEUROSCI.17-15-05843.1997
Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits
Abstract
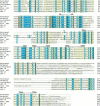
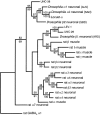
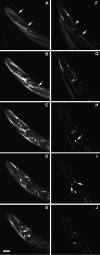
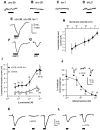
We show that three of the eleven genes of the nematode Caenorhabditis elegans that mediate resistance to the nematocide levamisole and to other cholinergic agonists encode nicotinic acetylcholine receptor (nAChR) subunits. unc-38 encodes an alpha subunit while lev-1 and unc-29 encode non-alpha subunits. The nematode nAChR subunits show conservation of many mammalian nAChR sequence features, implying an ancient evolutionary origin of nAChR proteins. Expression in Xenopus oocytes of combinations of these subunits that include the unc-38 alpha subunit results in levamisole-induced currents that are suppressed by the nAChR antagonists mecamylamine, neosurugatoxin, and d-tubocurarine but not alpha-bungarotoxin. The mutant phenotypes reveal that unc-38 and unc-29 subunits are necessary for nAChR function, whereas the lev-1 subunit is not. An UNC-29-GFP fusion shows that UNC-29 is expressed in body and head muscles. Two dominant mutations of lev-1 result in a single amino acid substitution or addition in or near transmembrane domain 2, a region important to ion channel conductance and desensitization. The identification of viable nAChR mutants in C. elegans provides an advantageous system in which receptor expression and synaptic targeting can be manipulated and studied in vivo.
Figures



References
-
- Abramson SN, Culver P, Kline T, Li Y, Guest P, Gutman L, Taylor P. Lophotoxin and related coral toxins covalently label the α-subunit of the nicotinic acetylcholine receptor. J Biol Chem. 1988;263:18568–18573. - PubMed
-
- Abramson SN, Li Y, Culver P, Taylor P. An analog of lophotoxin reacts covalently with Tyr190 in the α-subunit of the nicotinic acetylcholine receptor. J Biol Chem. 1989;264:12666–12672. - PubMed
-
- Ajuh PM, Egwang TG. Cloning of a cDNA encoding a putative nicotinic acetylcholine receptor subunit of the human filarial parasite Onchocerca volvulus. Gene. 1994;144:127–129. - PubMed
-
- Arpagaus M, Fedon Y, Cousin X, Chatonne A, Berge J-B, Fournier D, Toutant J-P. cDNA sequence, gene structure, and in vitro expression of ace-1, the gene encoding acetylcholinesterase of class A in the nematode Caenorhabditis elegans. J Biol Chem. 1994;269:9957–9965. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
