Transposable elements as sources of variation in animals and plants
- PMID: 9223252
- PMCID: PMC33680
- DOI: 10.1073/pnas.94.15.7704
Transposable elements as sources of variation in animals and plants
Abstract
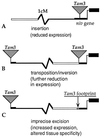
A tremendous wealth of data is accumulating on the variety and distribution of transposable elements (TEs) in natural populations. There is little doubt that TEs provide new genetic variation on a scale, and with a degree of sophistication, previously unimagined. There are many examples of mutations and other types of genetic variation associated with the activity of mobile elements. Mutant phenotypes range from subtle changes in tissue specificity to dramatic alterations in the development and organization of tissues and organs. Such changes can occur because of insertions in coding regions, but the more sophisticated TE-mediated changes are more often the result of insertions into 5' flanking regions and introns. Here, TE-induced variation is viewed from three evolutionary perspectives that are not mutually exclusive. First, variation resulting from the intrinsic parasitic nature of TE activity is examined. Second, we describe possible coadaptations between elements and their hosts that appear to have evolved because of selection to reduce the deleterious effects of new insertions on host fitness. Finally, some possible cases are explored in which the capacity of TEs to generate variation has been exploited by their hosts. The number of well documented cases in which element sequences appear to confer useful traits on the host, although small, is growing rapidly.
Figures
References
-
- McClintock B. Carnegie Inst Wash Yearbook. 1948;47:155–169.
-
- SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. - PubMed
-
- Finnegan D J. Curr Opin Genet Dev. 1992;2:861–867. - PubMed
-
- McClure M A. Reverse Transcriptase. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 425–443.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

