Vagaries of the molecular clock
- PMID: 9223263
- PMCID: PMC33703
- DOI: 10.1073/pnas.94.15.7776
Vagaries of the molecular clock
Abstract
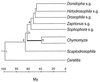
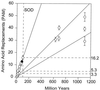
The hypothesis of the molecular evolutionary clock asserts that informational macromolecules (i.e., proteins and nucleic acids) evolve at rates that are constant through time and for different lineages. The clock hypothesis has been extremely powerful for determining evolutionary events of the remote past for which the fossil and other evidence is lacking or insufficient. I review the evolution of two genes, Gpdh and Sod. In fruit flies, the encoded glycerol-3-phosphate dehydrogenase (GPDH) protein evolves at a rate of 1.1 x 10(-10) amino acid replacements per site per year when Drosophila species are compared that diverged within the last 55 million years (My), but a much faster rate of approximately 4.5 x 10(-10) replacements per site per year when comparisons are made between mammals ( approximately 70 My) or Dipteran families ( approximately 100 My), animal phyla ( approximately 650 My), or multicellular kingdoms ( approximately 1100 My). The rate of superoxide dismutase (SOD) evolution is very fast between Drosophila species (16.2 x 10(-10) replacements per site per year) and remains the same between mammals (17.2) or Dipteran families (15.9), but it becomes much slower between animal phyla (5.3) and still slower between the three kingdoms (3.3). If we assume a molecular clock and use the Drosophila rate for estimating the divergence of remote organisms, GPDH yields estimates of 2,500 My for the divergence between the animal phyla (occurred approximately 650 My) and 3,990 My for the divergence of the kingdoms (occurred approximately 1,100 My). At the other extreme, SOD yields divergence times of 211 My and 224 My for the animal phyla and the kingdoms, respectively. It remains unsettled how often proteins evolve in such erratic fashion as GPDH and SOD.
Figures




References
-
- Zuckerkandl E, Pauling L. In: Horizons in Biochemistry. Kasha M, Pullman B, editors. New York: Academic; 1962. pp. 97–166.
-
- Zuckerkandl E, Pauling L. In: Evolving Genes and Proteins. Bryson V, Vogel H J, editors. New York: Academic; 1965. pp. 97–166. - PubMed
-
- Kimura M. Nature (London) 1968;217:624–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

