Evolution by the birth-and-death process in multigene families of the vertebrate immune system
- PMID: 9223266
- PMCID: PMC33709
- DOI: 10.1073/pnas.94.15.7799
Evolution by the birth-and-death process in multigene families of the vertebrate immune system
Abstract
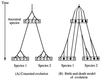
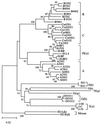
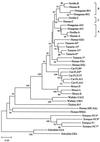
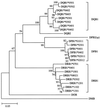
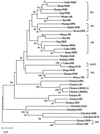
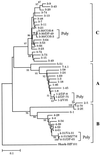
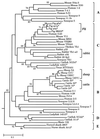
Concerted evolution is often invoked to explain the diversity and evolution of the multigene families of major histocompatibility complex (MHC) genes and immunoglobulin (Ig) genes. However, this hypothesis has been controversial because the member genes of these families from the same species are not necessarily more closely related to one another than to the genes from different species. To resolve this controversy, we conducted phylogenetic analyses of several multigene families of the MHC and Ig systems. The results show that the evolutionary pattern of these families is quite different from that of concerted evolution but is in agreement with the birth-and-death model of evolution in which new genes are created by repeated gene duplication and some duplicate genes are maintained in the genome for a long time but others are deleted or become nonfunctional by deleterious mutations. We found little evidence that interlocus gene conversion plays an important role in the evolution of MHC and Ig multigene families.
Figures

References
-
- Smith G P. Cold Spring Harbor Symp Quant Biol. 1973;38:507–513. - PubMed
-
- Irwin D M, Wilson A C. J Biol Chem. 1990;265:4944–4952. - PubMed
-
- Arnheim N. In: Evolution of Genes and Proteins. Nei M, Koehn R K, editors. Sunderland, MA: Sinauer; 1983. pp. 38–61.
-
- Weiss E H, Mellor A L, Golden L, Fahrner K, Simpson E, Hurst J, Flavell R A. Nature (London) 1983;301:671–674. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

