Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma
- PMID: 9223312
- PMCID: PMC21554
- DOI: 10.1073/pnas.94.15.8047
Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma
Abstract
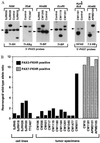
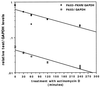
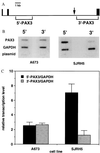
Chromosomal translocations identified in hematopoietic and solid tumors result in deregulated expression of protooncogenes or creation of chimeric proteins with tumorigenic potential. In the pediatric solid tumor alveolar rhabdomyosarcoma, a consistent t(2;13)(q35;q14) or variant t(1;13)(p36;q14) translocation generates PAX3-FKHR or PAX7-FKHR fusion proteins, respectively. In this report, we demonstrate that in addition to functional alterations these translocations are associated with fusion product overexpression. Furthermore, PAX3-FKHR and PAX7-FKHR overexpression occurs by distinct mechanisms. Transcription of PAX3-FKHR is increased relative to wild-type PAX3 by a copy number-independent process. In contrast, PAX7-FKHR overexpression results from fusion gene amplification. Thus, gene-specific mechanisms were selected to overexpress PAX3-FKHR and PAX7-FKHR in alveolar rhabdomyosarcoma, presumably due to differences in regulation between the wild-type loci. We postulate that these overexpression mechanisms ensure a critical level of gene product for the oncogenic effects of these fusions.
Figures

References
-
- Rabbitts T H. Nature (London) 1994;372:143–149. - PubMed
-
- Whang-Peng J, Knutsen T, Theil K, Horowitz M E, Triche T. Genes Chromosomes Cancer. 1992;5:299–310. - PubMed
-
- Biegel J A, Meek R S, Parmiter A H, Conard K, Emanuel B S. Genes Chromosomes Cancer. 1991;3:483–484. - PubMed
-
- Barr F G, Galili N, Holick J, Biegel J A, Rovera G, Emanuel B S. Nat Genet. 1993;3:113–117. - PubMed
-
- Galili N, Davis R J, Fredericks W J, Mukhopadhyay S, Rauscher F J, III, Emanuel B S, Rovera G, Barr F G. Nat Genet. 1993;5:230–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

