Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity
- PMID: 9230067
- PMCID: PMC2138183
- DOI: 10.1083/jcb.138.2.225
Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity
Abstract
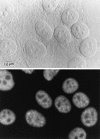
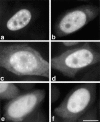
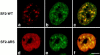
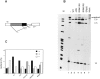
SR proteins are required for constitutive pre-mRNA splicing and also regulate alternative splice site selection in a concentration-dependent manner. They have a modular structure that consists of one or two RNA-recognition motifs (RRMs) and a COOH-terminal arginine/serine-rich domain (RS domain). We have analyzed the role of the individual domains of these closely related proteins in cellular distribution, subnuclear localization, and regulation of alternative splicing in vivo. We observed striking differences in the localization signals present in several human SR proteins. In contrast to earlier studies of RS domains in the Drosophila suppressor-of-white-apricot (SWAP) and Transformer (Tra) alternative splicing factors, we found that the RS domain of SF2/ASF is neither necessary nor sufficient for targeting to the nuclear speckles. Although this RS domain is a nuclear localization signal, subnuclear targeting to the speckles requires at least two of the three constituent domains of SF2/ASF, which contain additive and redundant signals. In contrast, in two SR proteins that have a single RRM (SC35 and SRp20), the RS domain is both necessary and sufficient as a targeting signal to the speckles. We also show that RRM2 of SF2/ASF plays an important role in alternative splicing specificity: deletion of this domain results in a protein that, although active in alternative splicing, has altered specificity in 5' splice site selection. These results demonstrate the modularity of SR proteins and the importance of individual domains for their cellular localization and alternative splicing function in vivo.
Figures







References
-
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. - PubMed
-
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. - PubMed
-
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

