Detection of functional nicotinic receptors blocked by alpha-bungarotoxin on PC12 cells and dependence of their expression on post-translational events
- PMID: 9236221
- PMCID: PMC6568351
- DOI: 10.1523/JNEUROSCI.17-16-06094.1997
Detection of functional nicotinic receptors blocked by alpha-bungarotoxin on PC12 cells and dependence of their expression on post-translational events
Abstract
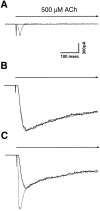
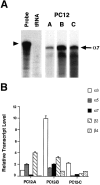
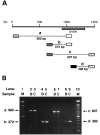
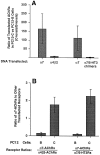
A major class of nicotinic receptors in the nervous system is one that binds alpha-bungarotoxin and contains the alpha7 gene product. PC12 cells, frequently used to study nicotinic receptors, express the alpha7 gene and have binding sites for the toxin, but previous attempts to elicit currents from the putative receptors have failed. Using whole-cell patch-clamp recording techniques and rapid application of agonist, we find a rapidly desensitizing acetylcholine-induced current in the cells that can be blocked by alpha-bungarotoxin. The current amplitude varies dramatically among three populations of PC12 cells but correlates well with the number of toxin-binding receptors. In contrast, the current shows no correlation with alpha7 transcript; cells with high levels of alpha7 mRNA can be negative for toxin binding and yet have other functional nicotinic receptors. Northern blot analysis and reverse transcription-PCR reveal no defects in alpha7 RNA from the negative cells, and immunoblot analysis demonstrates that they contain full-length alpha7 protein, although at reduced levels. Affinity purification of toxin-binding receptors from cells expressing them confirms that the receptors contain alpha7 protein. Transfection experiments demonstrate that PC12 cells lacking native toxin-binding receptors are deficient at producing receptors from alpha7 gene constructs, although the same cells can produce receptors from other transfected gene constructs. The results indicate that nicotinic receptors that bind alpha-bungarotoxin and contain alpha7 subunits require additional gene products to facilitate assembly and stabilization of the receptors. PC12 cells offer a model system for identifying those gene products.
Figures



References
-
- Alkondon M, Albuquerque EX. Diversity of nicotinic acetylcholine receptors in rat hippocampal neurons. I. Pharmacological and functional evidence for distinct structural subtypes. J Pharmacol Exp Ther. 1993;265:1455–1473. - PubMed
-
- Alkondon M, Reinhardt S, Lobron C, Hermsen B, Maelicke A, Albuquerque EX. Diversity of nicotinic acetylcholine receptors in rat hippocampal neurons. II. The rundown and inward rectification of agonist-elicited whole-cell currents and identification of receptor subunits by in situ hybridization. J Pharmacol Exp Ther. 1994;271:494–506. - PubMed
-
- Anand R, Peng X, Lindstrom J. Homomeric and native α7 acetylcholine receptors exhibit remarkably similar but non-identical pharmacological properties, suggesting that the native receptor is a heteromeric protein complex. FEBS Lett. 1993;327:241–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
