Philopatry of male marine turtles inferred from mitochondrial DNA markers
- PMID: 9238077
- PMCID: PMC23194
- DOI: 10.1073/pnas.94.16.8912
Philopatry of male marine turtles inferred from mitochondrial DNA markers
Abstract
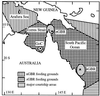
Recent studies of mitochondrial DNA (mtDNA) variation among marine turtle populations are consistent with the hypothesis that females return to beaches in their natal region to nest as adults. In contrast, less is known about breeding migrations of male marine turtles and whether they too are philopatric to natal regions. Studies of geographic structuring of restriction fragment and microsatellite polymorphisms at anonymous nuclear loci in green turtle (Chelonia mydas) populations indicate that nuclear gene flow is higher than estimates from mtDNA analyses. Regional populations from the northern and southern Great Barrier Reef were distinct for mtDNA but indistinguishable at nuclear loci, whereas the Gulf of Carpentaria (northern Australia) population was distinct for both types of marker. To assess whether this result was due to reduced philopatry of males across the Great Barrier Reef, we determined the mtDNA haplotypes of breeding males at courtship areas for comparison with breeding females from the same three locations. We used a PCR-restriction fragment length polymorphism approach to determine control region haplotypes and designed mismatch primers for the identification of specific haplotypes. The mtDNA haplotype frequencies were not significantly different between males and females at any of the three areas and estimates of Fst among the regions were similar for males and females (Fst = 0.78 and 0.73, respectively). We conclude that breeding males, like females, are philopatric to courtship areas within their natal region. Nuclear gene flow between populations is most likely occurring through matings during migrations of both males and females through nonnatal courtship areas.
Figures

Similar articles
-
Global population genetic structure and male-mediated gene flow in the green turtle (Chelonia mydas): RFLP analyses of anonymous nuclear loci.Genetics. 1992 May;131(1):163-73. doi: 10.1093/genetics/131.1.163. Genetics. 1992. PMID: 1350555 Free PMC article.
-
Geographic structure of mitochondrial and nuclear gene polymorphisms in Australian green turtle populations and male-biased gene flow.Genetics. 1997 Dec;147(4):1843-54. doi: 10.1093/genetics/147.4.1843. Genetics. 1997. PMID: 9409840 Free PMC article.
-
Mitochondrial DNA control region polymorphisms: genetic markers for ecological studies of marine turtles.Mol Ecol. 1994 Aug;3(4):363-73. doi: 10.1111/j.1365-294x.1994.tb00076.x. Mol Ecol. 1994. PMID: 7921361
-
Population genetics and phylogeography of sea turtles.Mol Ecol. 2007 Dec;16(23):4886-907. doi: 10.1111/j.1365-294X.2007.03542.x. Epub 2007 Oct 16. Mol Ecol. 2007. PMID: 17944856
-
Investigating sea turtle migration using DNA markers.Curr Opin Genet Dev. 1994 Dec;4(6):882-6. doi: 10.1016/0959-437x(94)90074-4. Curr Opin Genet Dev. 1994. PMID: 7888759 Review.
Cited by
-
Demographic changes in Pleistocene sea turtles were driven by past sea level fluctuations affecting feeding habitat availability.Mol Ecol. 2022 Feb;31(4):1044-1056. doi: 10.1111/mec.16302. Epub 2021 Dec 14. Mol Ecol. 2022. PMID: 34861074 Free PMC article.
-
Strong male-biased operational sex ratio in a breeding population of loggerhead turtles (Caretta caretta) inferred by paternal genotype reconstruction analysis.Ecol Evol. 2013 Nov;3(14):4736-47. doi: 10.1002/ece3.761. Epub 2013 Oct 30. Ecol Evol. 2013. PMID: 24363901 Free PMC article.
-
Distribution of genetic diversity reveals colonization patterns and philopatry of the loggerhead sea turtles across geographic scales.Sci Rep. 2020 Oct 22;10(1):18001. doi: 10.1038/s41598-020-74141-6. Sci Rep. 2020. PMID: 33093463 Free PMC article.
-
Multiple distant origins for green sea turtles aggregating off Gorgona Island in the Colombian eastern Pacific.PLoS One. 2012;7(2):e31486. doi: 10.1371/journal.pone.0031486. Epub 2012 Feb 2. PLoS One. 2012. PMID: 22319635 Free PMC article.
-
The population genomic structure of green turtles (Chelonia mydas) suggests a warm-water corridor for tropical marine fauna between the Atlantic and Indian oceans during the last interglacial.Heredity (Edinb). 2021 Dec;127(6):510-521. doi: 10.1038/s41437-021-00475-0. Epub 2021 Oct 11. Heredity (Edinb). 2021. PMID: 34635850 Free PMC article.
References
-
- Carr A. Conserv Biol. 1987;1:103–121.
-
- Carr A. Copeia. 1975;1975:547–555.
-
- Limpus C J, Miller J D, Parmenter C J, Reimer D, McLachlan N, Webb R. Wildl Res. 1992;19:347–358.
-
- Hendrickson J R. Proc Zool Soc London. 1958;130:455–535.
-
- Carr A, Ogren L. Bull Am Mus Nat Hist. 1960;121:7–48.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

