Genotator: a workbench for sequence annotation
- PMID: 9253604
- PMCID: PMC310682
- DOI: 10.1101/gr.7.7.754
Genotator: a workbench for sequence annotation
Abstract
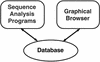
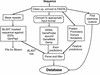
Sequencing centers such as the Human Genome Center at LBNL are producing an ever-increasing flood of genetic data. Annotation can greatly enhance the biological value of these sequences. Useful annotations include possible gene locations, homologies to known genes, and gene signal such as promoters and splice sites. Genotator is a workbench for automated sequence annotation and annotation browsing. The back end runs a series of sequence analysis tools on a DNA sequence, handling the various input and output formats required by the tools. Genotator currently runs five different gene-finding programs, three homology searches, and searches for promoters, splice sites, and ORFs. The results of the analyses run by Genotator can be viewed with the interactive graphical browser. The browser displays color-coded sequence annotations on a canvas that can be scrolled and zoomed, allowing the annotated sequence to be explored at multiple levels of detail. The user can view the actual DNA sequence in a separate window; when a region is selected in the map display, it is highlighted automatically in the sequence display, and vice versa. By displaying the output of all of the sequence analyses, Genotator provides an intuitive way to identify the significant regions (for example, probable exons) in a sequence. Users can interactively add personal annotations to label regions of interest. Additional capabilities of Genotator include primer design and pattern searching.
Figures





References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Borodovsky M, McIninch JD. GENEMARK: Parallel gene recognition for both DNA strands. Comput & Chem. 1993;17:123–133.
-
- Claverie JM, States DJ. Information enhancement methods for large scale sequence analysis. Comput & Chem. 1993;17:191–201.
-
- Durbin, R. and J. Thierry-Mieg. 1991. A C. elegans database. Documentation, code, and data available from anonymous FTP servers at lirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk and ncbi.nlm.nih.gov.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
