Evidence that the homeodomain protein Gtx is involved in the regulation of oligodendrocyte myelination
- PMID: 9254678
- PMCID: PMC6573154
- DOI: 10.1523/JNEUROSCI.17-17-06657.1997
Evidence that the homeodomain protein Gtx is involved in the regulation of oligodendrocyte myelination
Abstract
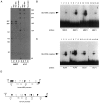
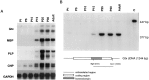
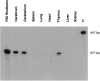
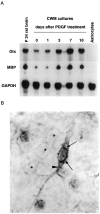
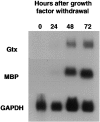
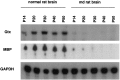
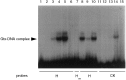
We have investigated the patterns of postnatal brain expression and DNA binding of Gtx, a homeodomain transcription factor. Gtx mRNA accumulates in parallel with the RNAs encoding the major structural proteins of myelin, myelin basic protein (MBP), and proteolipid protein (PLP) during postnatal brain development; Gtx mRNA decreases in parallel with MBP and PLP mRNAs in the brains of myelin-deficient rats, which have a point mutation in the PLP gene. Gtx mRNA is expressed in differentiated, postmitotic oligodendrocytes but is not found in oligodendrocyte precursors or astrocytes. These data thus demonstrate that Gtx is expressed uniquely in differentiated oligodendrocytes in postnatal rodent brain and that its expression is regulated in parallel with the major myelin protein mRNAs, encoding MBP and PLP, under a variety of physiologically relevant circumstances. Using a Gtx fusion protein produced in bacteria, we have confirmed that Gtx is a sequence-specific DNA-binding protein, which binds DNA sequences containing a core AT-rich homeodomain binding site. Immunoprecipitation of labeled DNA fragments encoding either the MBP or PLP promoter regions with this fusion protein has identified several Gtx-binding fragments, and we have confirmed these data using an electrophoretic mobility shift assay. In this way we have identified four Gtx binding sites within the first 750 bp of the MBP promoter and four Gtx binding sites within the first 1. 3 kb of the PLP promoter. In addition, inspection of the PLP promoter sequence demonstrates the presence of six additional Gtx binding sites. These data, taken together, strongly suggest that Gtx is important for the function of differentiated oligodendrocytes and may be involved in the regulation of myelin-specific gene expression.
Figures
Similar articles
-
The re-expression of the homeodomain transcription factor Gtx during remyelination of experimentally induced demyelinating lesions in young and old rat brain.Neuroscience. 2000;100(1):131-9. doi: 10.1016/s0306-4522(00)00252-9. Neuroscience. 2000. PMID: 10996464
-
Regulation of myelin development.Mult Scler. 1996 Dec;2(5):236-40. doi: 10.1177/135245859600200506. Mult Scler. 1996. PMID: 9050362 Review.
-
Gtx, an oligodendrocyte-specific homeodomain protein, has repressor activity.J Neurosci Res. 2000 Aug 15;61(4):376-87. doi: 10.1002/1097-4547(20000815)61:4<376::AID-JNR4>3.0.CO;2-#. J Neurosci Res. 2000. PMID: 10931524
-
Nuclear organization in differentiating oligodendrocytes.J Cell Sci. 2002 Nov 1;115(Pt 21):4071-9. doi: 10.1242/jcs.00103. J Cell Sci. 2002. PMID: 12356912
-
The pathobiology of myelin mutants reveal novel biological functions of the MBP and PLP genes.Brain Pathol. 2001 Jan;11(1):74-91. doi: 10.1111/j.1750-3639.2001.tb00383.x. Brain Pathol. 2001. PMID: 11145205 Free PMC article. Review.
Cited by
-
CNS myelin paranodes require Nkx6-2 homeoprotein transcriptional activity for normal structure.J Neurosci. 2004 Dec 15;24(50):11215-25. doi: 10.1523/JNEUROSCI.3479-04.2004. J Neurosci. 2004. PMID: 15601927 Free PMC article.
-
Enhancer Dynamics and Spatial Organization Drive Anatomically Restricted Cellular States in the Human Spinal Cord.bioRxiv [Preprint]. 2025 Jan 11:2025.01.10.632483. doi: 10.1101/2025.01.10.632483. bioRxiv. 2025. PMID: 39829819 Free PMC article. Preprint.
-
Co-localization of Nkx6.2 and Nkx2.2 homeodomain proteins in differentiated myelinating oligodendrocytes.Glia. 2010 Mar;58(4):458-68. doi: 10.1002/glia.20937. Glia. 2010. PMID: 19780200 Free PMC article.
-
rRNA-complementarity in the 5' untranslated region of mRNA specifying the Gtx homeodomain protein: evidence that base- pairing to 18S rRNA affects translational efficiency.Proc Natl Acad Sci U S A. 1999 Feb 16;96(4):1339-44. doi: 10.1073/pnas.96.4.1339. Proc Natl Acad Sci U S A. 1999. PMID: 9990025 Free PMC article.
-
Mice lacking the Nkx6.2 (Gtx) homeodomain transcription factor develop and reproduce normally.Mol Cell Biol. 2001 Jul;21(13):4399-403. doi: 10.1128/MCB.21.13.4399-4403.2001. Mol Cell Biol. 2001. PMID: 11390667 Free PMC article.
References
-
- Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in PFP1/PDX1-deficient mice. Development. 1996;122:1409–1416. - PubMed
-
- Armstrong RC, Kim JG, Hudson LD. Expression of myelin transcription factor I (MyTI), a “zinc-finger” DNA-Binding protein, in developing oligodendrocytes. Glia. 1995;14:303–321. - PubMed
-
- Arroyo EJ, Wegener M, Rosenfeld MG, Kamholz J, Scherer SS. Oligodendrocyte precursors express Tst-1 (SCIP/Oct-6) in vivo. Soc Neurosci Abstr. 1994;20:456.
-
- Berndt JA, Kim JG, Hudson LD. Identification of cis-regulatory elements in the myelin proteolipid protein (PLP) gene. J Biol Chem. 1992;267:14730–14737. - PubMed
-
- Bernier L, Alvarez F, Norgard EM, Raible DW, Mentaberry A, Schembri JG, Sabatini DD, Colman DR. Molecular cloning of a 2′,3′-cyclic nucleotide 3′-phosphodiesterase: mRNAs with different 5′ ends encode the same set of proteins in nervous and lymphoid tissues. J Neurosci. 1987;7:2703–2710. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
