Three functional luciferase domains in a single polypeptide chain
- PMID: 9256416
- PMCID: PMC22980
- DOI: 10.1073/pnas.94.17.8954
Three functional luciferase domains in a single polypeptide chain
Abstract
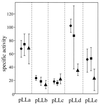
We report a unique case of a gene containing three homologous and contiguous repeat sequences, each of which, after excision, cloning, and expression in Escherichia coli, is shown to code for a peptide catalyzing the same reaction as the native protein, Gonyaulax polyedra luciferase (Mr = 137). This enzyme, which catalyzes the light-emitting oxidation of a linear tetrapyrrole (dinoflagellate luciferin), exhibits no sequence similarities to other luciferases in databases. Sequence analysis also reveals an unusual evolutionary feature of this gene: synonymous substitutions are strongly constrained in the central regions of each of the repeated coding sequences.
Figures



References
-
- Krasnow R, Dunlap J, Taylor W, Hastings J W, Vetterling W, Gooch V D. J Comp Physiol. 1980;138:19–26.
-
- Nicolas M-T, Morse D, Bassot J-M, Hastings J W. Protoplasma. 1991;160:159–166.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

