Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane
- PMID: 9256512
- PMCID: PMC23240
- DOI: 10.1073/pnas.94.17.9504
Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane
Abstract
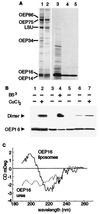
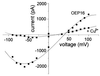
The reconstituted pea chloroplastic outer envelope protein of 16 kDa (OEP16) forms a slightly cation-selective, high-conductance channel with a conductance of Lambda = 1,2 nS (in 1 M KCl). The open probability of OEP16 channel is highest at 0 mV (Popen = 0.8), decreasing exponentially with higher potentials. Transport studies using reconstituted recombinant OEP16 protein show that the OEP16 channel is selective for amino acids but excludes triosephosphates or uncharged sugars. Crosslinking indicates that OEP16 forms a homodimer in the membrane. According to its primary sequence and predicted secondary structure, OEP16 shows neither sequence nor structural homologies to classical porins. The results indicate that the intermembrane space between the two envelope membranes might not be as freely accessible as previously thought.
Figures


References
-
- Gray J C, Row P E. Trends Cell Biol. 1995;5:243–251. - PubMed
-
- Schnell D J. Cell. 1995;83:521–524. - PubMed
-
- Soll J. Bot Acta. 1995;108:277–282.
-
- Joyard J, Block M A, Douce R. Eur J Biochem. 1991;199:489–509. - PubMed
-
- Lam H-M, Coschigano K T, Oliveira I C, Melo-Oliveira R, Coruzzi G M. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:569–593. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

