The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo
- PMID: 9275166
- PMCID: PMC23225
- DOI: 10.1073/pnas.94.18.9585
The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo
Abstract
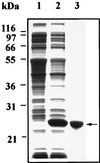
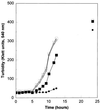
A gene homologous to methionine sulfoxide reductase (msrA) was identified as the predicted ORF (cosmid 9379) in chromosome V of Saccharomyces cerevisiae encoding a protein of 184 amino acids. The corresponding protein has been expressed in Escherichia coli and purified to homogeneity. The recombinant yeast MsrA possessed the same substrate specificity as the other known MsrA enzymes from mammalian and bacterial cells. Interruption of the yeast gene resulted in a null mutant, DeltamsrA::URA3 strain, which totally lost its cellular MsrA activity and was shown to be more sensitive to oxidative stress in comparison to its wild-type parent strain. Furthermore, high levels of free and protein-bound methionine sulfoxide were detected in extracts of msrA mutant cells relative to their wild-type parent cells, under various oxidative stresses. These findings show that MsrA is responsible for the reduction of methionine sulfoxide in vivo as well as in vitro in eukaryotic cells. Also, the results support the proposition that MsrA possess an antioxidant function. The ability of MsrA to repair oxidative damage in vivo may be of singular importance if methionine residues serve as antioxidants.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

