A novel virus in swine is closely related to the human hepatitis E virus
- PMID: 9275216
- PMCID: PMC23282
- DOI: 10.1073/pnas.94.18.9860
A novel virus in swine is closely related to the human hepatitis E virus
Abstract
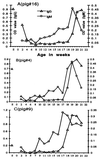
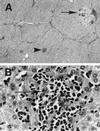
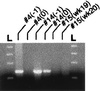
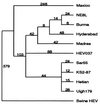
A novel virus, designated swine hepatitis E virus (swine HEV), was identified in pigs. Swine HEV crossreacts with antibody to the human HEV capsid antigen. Swine HEV is a ubiquitous agent and the majority of swine >/=3 months of age in herds from the midwestern United States were seropositive. Young pigs naturally infected by swine HEV were clinically normal but had microscopic evidence of hepatitis, and developed viremia prior to seroconversion. The entire ORFs 2 and 3 were amplified by reverse transcription-PCR from sera of naturally infected pigs. The putative capsid gene (ORF2) of swine HEV shared about 79-80% sequence identity at the nucleotide level and 90-92% identity at the amino acid level with human HEV strains. The small ORF3 of swine HEV had 83-85% nucleotide sequence identity and 77-82% amino acid identity with human HEV strains. Phylogenetic analyses showed that swine HEV is closely related to, but distinct from, human HEV strains. The discovery of swine HEV not only has implications for HEV vaccine development, diagnosis, and biology, but also raises a potential public health concern for zoonosis or xenozoonosis following xenotransplantation with pig organs.
Figures

References
-
- Purcell R H. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott–Raven; 1996. pp. 2831–2843.
-
- Wong D C, Purcell R H, Sreenivasan M A, Prasad S R, Pavri K M. Lancet. 1980;ii:876–878. - PubMed
-
- Arankalle V A, Tsarev S A, Chadha M S, Alling D W, Emerson S U, Banerjee K, Purcell R H. J Infect Dis. 1995;171:447–450. - PubMed
-
- Bradley D W. Rev Med Virol. 1992;2:19–28.
-
- Skidmore S J, Yarbough P O, Gabor K A, Tam A W, Reyes G R. Lancet. 1991;337:1541. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

