Exploring the folding free energy surface of a three-helix bundle protein
- PMID: 9294180
- PMCID: PMC23332
- DOI: 10.1073/pnas.94.19.10161
Exploring the folding free energy surface of a three-helix bundle protein
Abstract
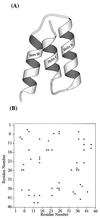
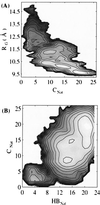
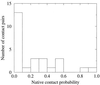
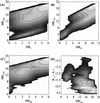
The multidimensional free energy surface for a small fast folding helical protein is explored based on first-principle calculations. The model represents the 46-residue segment from fragment B of staphylococcal protein A. The relationship between collapse and tertiary structure formation, and the order of collapse and secondary structure formation, are investigated. We find that the initial collapse process gives rise to a transition state with about 30% of the native tertiary structure and 50-70% of the native helix content. We also observe two distinct distributions of native helix in this collapsed state (Rg approximately 12 A), one with about 20% of the native helical hydrogen bonds, the other with near 70%. The former corresponds to a local minimum. The barrier from this metastable state to the native state is about 2 kBT. In the latter case, folding is essentially a downhill process involving topological assembly. In addition, the order of formation of secondary structure among the three helices is examined. We observe cooperative formation of the secondary structure in helix I and helix II. Secondary structure in helix III starts to form following the formation of certain secondary structure in both helix I and helix II. Comparisons of our results with those from theory and experiment are made.
Figures
References
-
- Kim P S, Baldwin R L. Annu Rev Biochem. 1990;59:631–660. - PubMed
-
- Karplus M, Weaver D L. Biopolymers. 1979;18:1421–1437.
-
- Ptitsyn O B. J Protein Chem. 1987;6:273–293.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

