A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans
- PMID: 9294229
- PMCID: PMC23381
- DOI: 10.1073/pnas.94.19.10438
A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans
Abstract
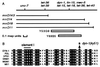
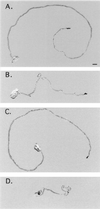
Drosophila shibire and its mammalian homologue dynamin regulate an early step in endocytosis. We identified a Caenorhabditis elegans dynamin gene, dyn-1, based upon hybridization to the Drosophila gene. The dyn-1 RNA transcripts are trans-spliced to the spliced leader 1 and undergo alternative splicing to code for either an 830- or 838-amino acid protein. These dyn-1 proteins are highly similar in amino acid sequence, structure, and size to the Drosophila and mammalian dynamins: they contain an N-terminal GTPase, a pleckstrin homology domain, and a C-terminal proline-rich domain. We isolated a recessive temperature-sensitive dyn-1 mutant containing an alteration within the GTPase domain that becomes uncoordinated when shifted to high temperature and that recovers when returned to lower temperatures, similar to D. shibire mutants. When maintained at higher temperatures, dyn-1 mutants become constipated, egg-laying defective, and produce progeny that die during embryogenesis. Using a dyn-1::lacZ gene fusion, a high level of dynamin expression was observed in motor neurons, intestine, and pharyngeal muscle. Our results suggest that dyn-1 function is required during development and for normal locomotion.
Figures




References
-
- Baba T, Damke H, Hinshaw J, Ikeda K, Schmid S L, Warnock D E. Cold Spring Harbor Symp Quant Biol. 1995;60:235–242. - PubMed
-
- Shpetner H S, Vallee R B. Cell. 1989;59:421–432. - PubMed
-
- Chen M S, Obar R A, Schroeder C C, Austin T W, Poodry C A, Wadsworth S C, Vallee R B. Nature (London) 1991;351:583–586. - PubMed
-
- van der Bliek A M, Meyerowitz E M. Nature (London) 1991;351:411–414. - PubMed
-
- Grigliatti T A, Hall L, Rosenbluth R, Suzuki D T. Mol Gen Genet. 1973;120:107–114. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

