The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila
- PMID: 9295388
- PMCID: PMC6573436
- DOI: 10.1523/JNEUROSCI.17-19-07425.1997
The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila
Abstract
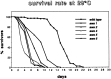
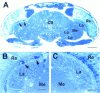
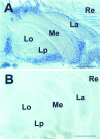
Swiss cheese (sws) mutant flies develop normally during larval life but show age-dependent neurodegeneration in the pupa and adult and have reduced life span. In late pupae, glial processes form abnormal, multilayered wrappings around neurons and axons. Degeneration first becomes evident in young flies as apoptosis in single scattered cells in the CNS, but later it becomes severe and widespread. In the adult, the number of glial wrappings increases with age. The sws gene is expressed in neurons in the brain cortex. The conceptual 1425 amino acid protein shows two domains with homology to the regulatory subunits of protein kinase A and to conceptual proteins of yet unknown function in yeast, worm, and human. Sequencing of two sws alleles shows amino acid substitutions in these two conserved domains. It is suggested that the novel SWS protein plays a role in a signaling mechanism between neurons and glia that regulates glial wrapping during development of the adult brain.
Figures





References
-
- Altschul S, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989.
-
- Barres BA, Raff MC. Control of oligodendrocyte number in the developing rat optic nerve. Neuron. 1994;12:935–942. - PubMed
-
- Bergoffen J, Scherer SS, Wang S, Scott MO, Bone LJ, Paul DL, Chen K, Lensch MW, Chance PF, Fischbeck KH. Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science. 1993;262:2039–2042. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
