Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis
- PMID: 9298992
- PMCID: PMC2132555
- DOI: 10.1083/jcb.138.6.1379
Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis
Abstract
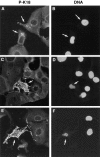
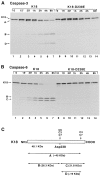
Keratins 8 (K8) and 18 (K18) are major components of intermediate filaments (IFs) of simple epithelial cells and tumors derived from such cells. Structural cell changes during apoptosis are mediated by proteases of the caspase family. During apoptosis, K18 IFs reorganize into granular structures enriched for K18 phosphorylated on serine 53. K18, but not K8, generates a proteolytic fragment during drug- and UV light-induced apoptosis; this fragment comigrates with K18 cleaved in vitro by caspase-6, -3, and -7. K18 is cleaved by caspase-6 into NH2-terminal, 26-kD and COOH-terminal, 22-kD fragments; caspase-3 and -7 additionally cleave the 22-kD fragment into a 19-kD fragment. The cleavage site common for the three caspases was the sequence VEVD/A, located in the conserved L1-2 linker region of K18. The additional site for caspases-3 and -7 that is not cleaved efficiently by caspase-6 is located in the COOH-terminal tail domain of K18. Expression of K18 with alanine instead of serine at position 53 demonstrated that cleavage during apoptosis does not require phosphorylation of serine 53. However, K18 with a glutamate instead of aspartate at position 238 was resistant to proteolysis during apoptosis. Furthermore, this cleavage site mutant appears to cause keratin filament reorganization in stably transfected clones. The identification of the L1-2 caspase cleavage site, and the conservation of the same or very similar sites in multiple other intermediate filament proteins, suggests that the processing of IFs during apoptosis may be initiated by a similar caspase cleavage.
Figures









References
-
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. - PubMed
-
- Anderson JM, Heindl LM, Bauman PA, Ludi CW, Dalton WS, Cress AE. Cytokeratin expression results in a drug-resistant phenotype to six different chemotherapeutic agents. Clin Cancer Res. 1996;2:97–105. - PubMed
-
- Baribault H, Price J, Miyai K, Oshima RG. Mid-gestational lethality in mice lacking keratin 8. Genes Dev. 1993;7:1191–1202. - PubMed
-
- Baribault H, Penner J, Iozzo RV, Wilson-Heiner M. Colorectal hyperplasia and inflammation in keratin 8-deficient FVB/N mice. Genes Dev. 1994;8:2964–2973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

