Translational repression by a transcriptional elongation factor
- PMID: 9303536
- PMCID: PMC275398
- DOI: 10.1101/gad.11.17.2204
Translational repression by a transcriptional elongation factor
Abstract
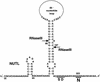
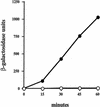
One of the classical positive regulators of gene expression is bacteriophage lambda N protein. N regulates the transcription of early phage genes by participating in the formation of a highly processive, terminator-resistant transcription complex and thereby stimulates the expression of genes lying downstream of transcriptional terminators. Also included in this antiterminating transcription complex are an RNA site (NUT) and host proteins (Nus). Here we demonstrate that N has an additional, hitherto unknown regulatory role, as a repressor of the translation of its own gene. N-dependent repression does not occur when NUT is deleted, demonstrating that N-mediated antitermination and translational repression both require the same cis-acting site in the RNA. In addition, we have identified one nut and several host mutations that eliminate antitermination and not translational repression, suggesting the independence of these two N-mediated mechanisms. Finally, the position of nutL with respect to the gene whose expression is repressed is important.
Figures
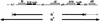



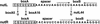
Similar articles
-
Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination.J Mol Biol. 1994 Feb 11;236(1):217-28. doi: 10.1006/jmbi.1994.1131. J Mol Biol. 1994. PMID: 8107107
-
Action of an RNA site at a distance: role of the nut genetic signal in transcription antitermination by phage-lambda N gene product.New Biol. 1990 Nov;2(11):975-91. New Biol. 1990. PMID: 2151659
-
The nut site of bacteriophage lambda is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase.Genes Dev. 1991 Nov;5(11):2141-51. doi: 10.1101/gad.5.11.2141. Genes Dev. 1991. PMID: 1834523
-
Genetic analysis of the N transcription antitermination system of phage lambda.Genome. 1989;31(2):491-6. doi: 10.1139/g89-096. Genome. 1989. PMID: 2534385 Review.
-
Transcription antitermination: the lambda paradigm updated.Mol Microbiol. 1995 Oct;18(2):191-200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. Mol Microbiol. 1995. PMID: 8709839 Review.
Cited by
-
The global regulator RNase III modulates translation repression by the transcription elongation factor N.EMBO J. 2002 Aug 1;21(15):4154-61. doi: 10.1093/emboj/cdf395. EMBO J. 2002. PMID: 12145215 Free PMC article.
-
RNase III: Genetics and function; structure and mechanism.Annu Rev Genet. 2013;47:405-31. doi: 10.1146/annurev-genet-110711-155618. Annu Rev Genet. 2013. PMID: 24274754 Free PMC article. Review.
-
Role of an RNase III binding site in transcription termination at lambda nutL by HK022 Nun protein.J Bacteriol. 2006 Oct;188(19):6824-31. doi: 10.1128/JB.00567-06. J Bacteriol. 2006. PMID: 16980485 Free PMC article.
-
Bacillus subtilis RNase III gene: cloning, function of the gene in Escherichia coli, and construction of Bacillus subtilis strains with altered rnc loci.J Bacteriol. 1997 Dec;179(23):7379-85. doi: 10.1128/jb.179.23.7379-7385.1997. J Bacteriol. 1997. PMID: 9393702 Free PMC article.
-
An efficient recombination system for chromosome engineering in Escherichia coli.Proc Natl Acad Sci U S A. 2000 May 23;97(11):5978-83. doi: 10.1073/pnas.100127597. Proc Natl Acad Sci U S A. 2000. PMID: 10811905 Free PMC article.
References
-
- Adhya S, Gottesman M, Court D. Independence of gene N and tof functions of bacteriophage lambda. J Mol Biol. 1977;112:657–660. - PubMed
-
- Barik S, Ghosh B, Whalen W, Lazinski D, Das A. An antitermination protein engages the elongating transcription apparatus at a promoter–proximal recognition site. Cell. 1987;50:885–899. - PubMed
-
- Braddock M, Chambers A, Wilson W, Esnouf MP, Adams SE, Kingsman AJ, Kingsman SM. HIV-1 TAT “activates” presynthesized RNA in the nucleus. Cell. 1989;58:269–279. - PubMed
-
- Braddock M, Thorburn AM, Chambers A, Elliot GD, Anderson GJ, Kingsman AJ, Kingsman SM. A nuclear translational block imposed by the HIV-1 U3 region is relieved by the Tat–TAR interaction. Cell. 1990;62:1123–1133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
